Characterization of a Bvg-regulated fatty acid methyl-transferase in Bordetella pertussis
- PMID: 28493897
- PMCID: PMC5426589
- DOI: 10.1371/journal.pone.0176396
Characterization of a Bvg-regulated fatty acid methyl-transferase in Bordetella pertussis
Abstract
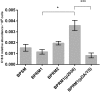
The whooping cough agent Bordetella pertussis controls the expression of its large virulence regulon in a coordinated manner through the two-component signal transduction system BvgAS. In addition to the genes coding for bona fide virulence factors, the Bvg regulon comprises genes of unknown function. In this work, we characterized a new Bvg-activated gene called BP2936. Homologs of BP2936 are found in other pathogenic Bordetellae and in several other species, including plant pathogens and environmental bacteria. We showed that the gene product of BP2936 is a membrane-associated methyl-transferase of free fatty acids. We thus propose to name it FmtB, for fatty acid methyl-transferase of Bordetella. The role of this protein was tested in cellular and animal models of infection, but the loss of BP2936 did not appear to affect host-pathogen interactions in those assays. The high level of conservation of BP2936 among B. pertussis isolates nevertheless argues that it probably plays a role in the life cycle of this pathogen.
Conflict of interest statement
Figures

References
-
- Melvin JA, Scheller EV, Miller JF, Cotter PA. Bordetella pertussis pathogenesis: current and future challenges. Nat Rev Microbiol. 2014; 12: 274–288. doi: 10.1038/nrmicro3235 - DOI - PMC - PubMed
-
- Libster Edwards CA. Re-emergence of pertussis: what are the solutions? Expert Rev Vaccines. 2012; 11: 1331–1346. doi: 10.1586/erv.12.118 - DOI - PubMed
-
- Rohani P, Zhong X, King AA. Contact network structure explains the changing epidemiology of pertussis. Science. 2010; 330: 982–985. doi: 10.1126/science.1194134 - DOI - PubMed
-
- Lavine JS, King AA, Bjornstad ON. Natural immune boosting in pertussis dynamics and the potential for long-term vaccine failure. Proc Natl Acad Sci U S A. 2011; 108: 7259–7264. doi: 10.1073/pnas.1014394108 - DOI - PMC - PubMed
-
- Cotter PA, Jones AM. Phosphorelay control of virulence gene expression in Bordetella. Trends Microbiol. 2003; 11: 367–373. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

