Sequencing of small RNAs of the fern Pleopeltis minima (Polypodiaceae) offers insight into the evolution of the microrna repertoire in land plants
- PMID: 28494025
- PMCID: PMC5426797
- DOI: 10.1371/journal.pone.0177573
Sequencing of small RNAs of the fern Pleopeltis minima (Polypodiaceae) offers insight into the evolution of the microrna repertoire in land plants
Abstract
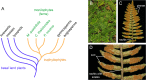
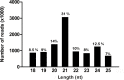
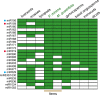
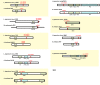
MicroRNAs (miRNAs) are short, single stranded RNA molecules that regulate the stability and translation of messenger RNAs in diverse eukaryotic groups. Several miRNA genes are of ancient origin and have been maintained in the genomes of animal and plant taxa for hundreds of millions of years, playing key roles in development and physiology. In the last decade, genome and small RNA (sRNA) sequencing of several plant species have helped unveil the evolutionary history of land plants. Among these, the fern group (monilophytes) occupies a key phylogenetic position, as it represents the closest extant cousin taxon of seed plants, i.e. gymno- and angiosperms. However, in spite of their evolutionary, economic and ecological importance, no fern genome has been sequenced yet and few genomic resources are available for this group. Here, we sequenced the small RNA fraction of an epiphytic South American fern, Pleopeltis minima (Polypodiaceae), and compared it to plant miRNA databases, allowing for the identification of miRNA families that are shared by all land plants, shared by all vascular plants (tracheophytes) or shared by euphyllophytes (ferns and seed plants) only. Using the recently described transcriptome of another fern, Lygodium japonicum, we also estimated the degree of conservation of fern miRNA targets in relation to other plant groups. Our results pinpoint the origin of several miRNA families in the land plant evolutionary tree with more precision and are a resource for future genomic and functional studies of fern miRNAs.
Conflict of interest statement
Figures
Similar articles
-
Conservation and divergence of small RNA pathways and microRNAs in land plants.Genome Biol. 2017 Aug 23;18(1):158. doi: 10.1186/s13059-017-1291-2. Genome Biol. 2017. PMID: 28835265 Free PMC article.
-
Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes.BMC Evol Biol. 2009 Jun 11;9:130. doi: 10.1186/1471-2148-9-130. BMC Evol Biol. 2009. PMID: 19519899 Free PMC article.
-
Antiquity of microRNAs and their targets in land plants.Plant Cell. 2005 Jun;17(6):1658-73. doi: 10.1105/tpc.105.032185. Epub 2005 Apr 22. Plant Cell. 2005. PMID: 15849273 Free PMC article.
-
Trends and concepts in fern classification.Ann Bot. 2014 Mar;113(4):571-94. doi: 10.1093/aob/mct299. Epub 2014 Feb 13. Ann Bot. 2014. PMID: 24532607 Free PMC article. Review.
-
Conservation and divergence in plant microRNAs.Plant Mol Biol. 2012 Sep;80(1):3-16. doi: 10.1007/s11103-011-9829-2. Epub 2011 Oct 14. Plant Mol Biol. 2012. PMID: 21996939 Review.
Cited by
-
MicroRNAs: From Mechanism to Organism.Front Cell Dev Biol. 2020 Jun 3;8:409. doi: 10.3389/fcell.2020.00409. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32582699 Free PMC article. Review.
-
Far-Red Light-Induced Azolla filiculoides Symbiosis Sexual Reproduction: Responsive Transcripts of Symbiont Nostoc azollae Encode Transporters Whilst Those of the Fern Relate to the Angiosperm Floral Transition.Front Plant Sci. 2021 Aug 11;12:693039. doi: 10.3389/fpls.2021.693039. eCollection 2021. Front Plant Sci. 2021. PMID: 34456937 Free PMC article.
-
Genomic insights into genetic diploidization in the homosporous fern Adiantum nelumboides.Genome Biol Evol. 2022 Aug 10;14(8):evac127. doi: 10.1093/gbe/evac127. Online ahead of print. Genome Biol Evol. 2022. PMID: 35946426 Free PMC article.
-
Comparative transcriptome analysis, unfolding the pathways regulating the seed-size trait in cultivated lentil (Lens culinaris Medik.).Front Genet. 2022 Aug 10;13:942079. doi: 10.3389/fgene.2022.942079. eCollection 2022. Front Genet. 2022. PMID: 36035144 Free PMC article.
-
microRNA 166: an evolutionarily conserved stress biomarker in land plants targeting HD-ZIP family.Physiol Mol Biol Plants. 2021 Nov;27(11):2471-2485. doi: 10.1007/s12298-021-01096-x. Epub 2021 Nov 11. Physiol Mol Biol Plants. 2021. PMID: 34924705 Free PMC article. Review.
References
-
- Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature 1997; 389: 33–39.
-
- Becker B, Marin B. Streptophyte algae and the origin of embryophytes. Ann Bot. 2009; 103: 999–1004. doi: 10.1093/aob/mcp044 - DOI - PMC - PubMed
-
- Pires ND, Dolan L. Morphological evolution in land plants: new designs with old genes. Philos Trans R Soc Lond B Biol Sci. 2012; 367: 508–18. doi: 10.1098/rstb.2011.0252 - DOI - PMC - PubMed
-
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008; 319: 64–9. doi: 10.1126/science.1150646 - DOI - PubMed
-
- Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011; 332: 960–3. doi: 10.1126/science.1203810 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

