Characterization of miR-122-independent propagation of HCV
- PMID: 28494029
- PMCID: PMC5441651
- DOI: 10.1371/journal.ppat.1006374
Characterization of miR-122-independent propagation of HCV
Abstract
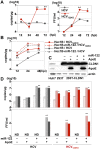
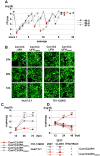
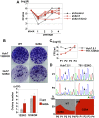
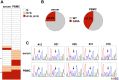
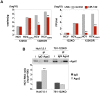
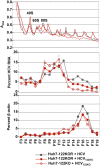
miR-122, a liver-specific microRNA, is one of the determinants for liver tropism of hepatitis C virus (HCV) infection. Although miR-122 is required for efficient propagation of HCV, we have previously shown that HCV replicates at a low rate in miR-122-deficient cells, suggesting that HCV-RNA is capable of propagating in an miR-122-independent manner. We herein investigated the roles of miR-122 in both the replication of HCV-RNA and the production of infectious particles by using miR-122-knockout Huh7 (Huh7-122KO) cells. A slight increase of intracellular HCV-RNA levels and infectious titers in the culture supernatants was observed in Huh7-122KO cells upon infection with HCV. Moreover, after serial passages of HCV in miR-122-knockout Huh7.5.1 cells, we obtained an adaptive mutant, HCV122KO, possessing G28A substitution in the 5'UTR of the HCV genotype 2a JFH1 genome, and this mutant may help to enhance replication complex formation, a possibility supported by polysome analysis. We also found the introduction of adaptive mutation around miR-122 binding site in the genotype 1b/2a chimeric virus, which originally had an adenine at the nucleotide position 29. HCV122KO exhibited efficient RNA replication in miR-122-knockout cells and non-hepatic cells without exogenous expression of miR-122. Competition assay revealed that the G28A mutant was dominant in the absence of miR-122, but its effects were equivalent to those of the wild type in the presence of miR-122, suggesting that the G28A mutation does not confer an advantage for propagation in miR-122-rich hepatocytes. These observations may explain the clinical finding that the positive rate of G28A mutation was higher in miR-122-deficient PBMCs than in the patient serum, which mainly included the hepatocyte-derived virus from HCV-genotype-2a patients. These results suggest that the emergence of HCV mutants that can propagate in non-hepatic cells in an miR-122-independent manner may participate in the induction of extrahepatic manifestations in chronic hepatitis C patients.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Galossi A, Guarisco R, Bellis L, Puoti C. Extrahepatic manifestations of chronic HCV infection. J Gastrointestin Liver Dis. 2007;16(1):65–73. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

