Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases
- PMID: 28494241
- PMCID: PMC5705050
- DOI: 10.1016/j.chom.2017.04.010
Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases
Abstract
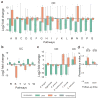
The gut microbiome plays a central role in inflammatory bowel diseases (IBDs) pathogenesis and propagation. To determine whether the gut microbiome may predict responses to IBD therapy, we conducted a prospective study with Crohn's disease (CD) or ulcerative colitis (UC) patients initiating anti-integrin therapy (vedolizumab). Disease activity and stool metagenomes at baseline, and weeks 14, 30, and 54 after therapy initiation were assessed. Community α-diversity was significantly higher, and Roseburia inulinivorans and a Burkholderiales species were more abundant at baseline among CD patients achieving week 14 remission. Several significant associations were identified with microbial function; 13 pathways including branched chain amino acid synthesis were significantly enriched in baseline samples from CD patients achieving remission. A neural network algorithm, vedoNet, incorporating microbiome and clinical data, provided highest classifying power for clinical remission. We hypothesize that the trajectory of early microbiome changes may be a marker of response to IBD treatment.
Keywords: Microbiome; butyrate; remission; roseburia; treatment response; vedolizumab.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


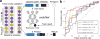
References
-
- Arijs I, Li K, Toedter G, Quintens R, Van Lommel L, Van Steen K, Leemans P, De Hertogh G, Lemaire K, Ferrante M, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58:1612–1619. - PubMed
-
- Baumgart DC, Sandborn WJ. Crohn's disease. Lancet. 2012;380:1590–1605. - PubMed
-
- Becker C, Neurath MF, Wirtz S. The Intestinal Microbiota in Inflammatory Bowel Disease. ILAR J. 2015;56:192–204. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

