Global Reprogramming of Host Kinase Signaling in Response to Fungal Infection
- PMID: 28494245
- PMCID: PMC5538893
- DOI: 10.1016/j.chom.2017.04.008
Global Reprogramming of Host Kinase Signaling in Response to Fungal Infection
Abstract
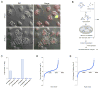
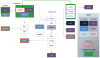
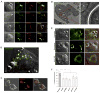
Cryptococcus neoformans (Cn) is a deadly fungal pathogen whose intracellular lifestyle is important for virulence. Host mechanisms controlling fungal phagocytosis and replication remain obscure. Here, we perform a global phosphoproteomic analysis of the host response to Cryptococcus infection. Our analysis reveals numerous and diverse host proteins that are differentially phosphorylated following fungal ingestion by macrophages, thereby indicating global reprogramming of host kinase signaling. Notably, phagocytosis of the pathogen activates the host autophagy initiation complex (AIC) and the upstream regulatory components LKB1 and AMPKα, which regulate autophagy induction through their kinase activities. Deletion of Prkaa1, the gene encoding AMPKα1, in monocytes results in resistance to fungal colonization of mice. Finally, the recruitment of AIC components to nascent Cryptococcus-containing vacuoles (CnCVs) regulates the intracellular trafficking and replication of the pathogen. These findings demonstrate that host AIC regulatory networks confer susceptibility to infection and establish a proteomic resource for elucidating host mechanisms that regulate fungal intracellular parasitism.
Keywords: AMPKα; Cryptococcus neoformans; autophagy initiation complex; fungal pathogen; host factors; host-pathogen interaction; kinase signaling; phagocytosis; phosphoproteomics; proteomics.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures



References
-
- Asara JM, Christofk HR, Freimark LM, Cantley LC. A label-free quantification method by MS/MS TIC compared to SILAC and spectral counting in a proteomics screen. Proteomics. 2008;8:994–999. - PubMed
-
- Bozzola JJ. Conventional specimen preparation techniques for transmission electron microscopy of cultured cells. Methods in molecular biology. 2007;369:1–18. - PubMed
-
- Chaki SP, Barhoumi R, Berginski ME, Sreenivasappa H, Trache A, Gomez SM, Rivera GM. Nck enables directional cell migration through the coordination of polarized membrane protrusion with adhesion dynamics. Journal of cell science. 2013;126:1637–1649. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

