Shotgun lipidomics in substantiating lipid peroxidation in redox biology: Methods and applications
- PMID: 28494428
- PMCID: PMC5423350
- DOI: 10.1016/j.redox.2017.04.030
Shotgun lipidomics in substantiating lipid peroxidation in redox biology: Methods and applications
Abstract
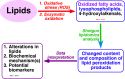
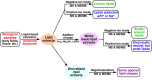
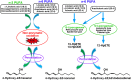
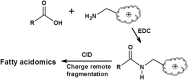
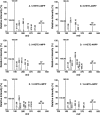
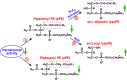
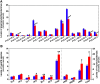
Multi-dimensional mass spectrometry-based shotgun lipidomics (MDMS-SL) has made profound advances for comprehensive analysis of cellular lipids. It represents one of the most powerful tools in analyzing lipids directly from lipid extracts of biological samples. It enables the analysis of nearly 50 lipid classes and thousands of individual lipid species with high accuracy/precision. The redox imbalance causes oxidative stress, resulting in lipid peroxidation, and alterations in lipid metabolism and homeostasis. Some lipid classes such as oxidized fatty acids, 4-hydroxyalkenal species, and plasmalogen are sensitive to oxidative stress or generated corresponding to redox imbalance. Therefore, accurate assessment of these lipid classes can provide not only the redox states, but also molecular insights into the pathogenesis of diseases. This review focuses on the advances of MDMS-SL in analysis of these lipid classes and molecular species, and summarizes their recent representative applications in biomedical/biological research. We believe that MDMS-SL can make great contributions to redox biology through substantiating the aberrant lipid metabolism, signaling, trafficking, and homeostasis under oxidative stress-related condition.
Keywords: 4-hydroxyalkenal species; Lipid peroxidation; Oxidative stress; Oxidized fatty acids; Plasmalogen; Shotgun lipidomics.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Wymann M.P., Schneiter R. Lipid signalling in disease. Nat. Rev. Mol. Cell Biol. 2008;9:162–176. - PubMed
-
- Mapstone M., Cheema A.K., Fiandaca M.S., Zhong X., Mhyre T.R., MacArthur L.H., Hall W.J., Fisher S.G., Peterson D.R., Haley J.M., Nazar M.D., Rich S.A., Berlau D.J., Peltz C.B., Tan M.T., Kawas C.H., Federoff H.J. Plasma phospholipids identify antecedent memory impairment in older adults. Nat. Med. 2014;20:415–418. - PMC - PubMed
-
- Han X. Lipid alterations in the earliest clinically recognizable stage of Alzheimer's disease: implication of the role of lipids in the pathogenesis of Alzheimer's disease. Curr. Alzheimer Res. 2005;2:65–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

