Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors
- PMID: 28494759
- PMCID: PMC5427549
- DOI: 10.1186/s12915-017-0379-1
Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors
Abstract
Background: Plant-pathogenic oomycetes are responsible for economically important losses in crops worldwide. Phytophthora palmivora, a tropical relative of the potato late blight pathogen, causes rotting diseases in many tropical crops including papaya, cocoa, oil palm, black pepper, rubber, coconut, durian, mango, cassava and citrus. Transcriptomics have helped to identify repertoires of host-translocated microbial effector proteins which counteract defenses and reprogram the host in support of infection. As such, these studies have helped in understanding how pathogens cause diseases. Despite the importance of P. palmivora diseases, genetic resources to allow for disease resistance breeding and identification of microbial effectors are scarce.
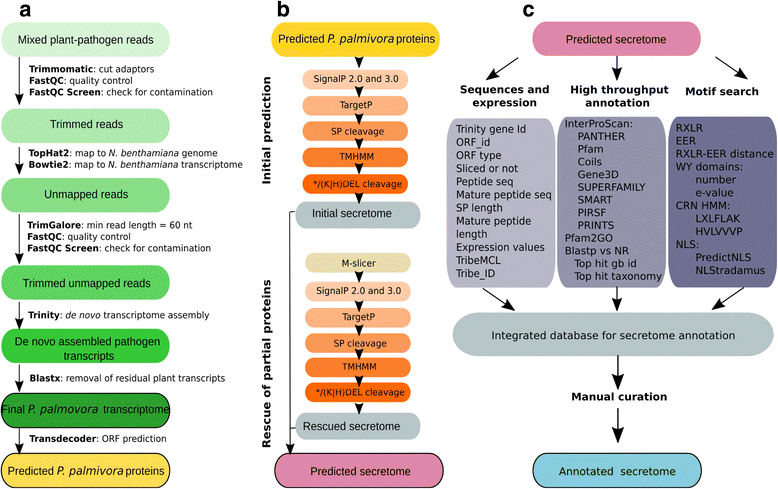
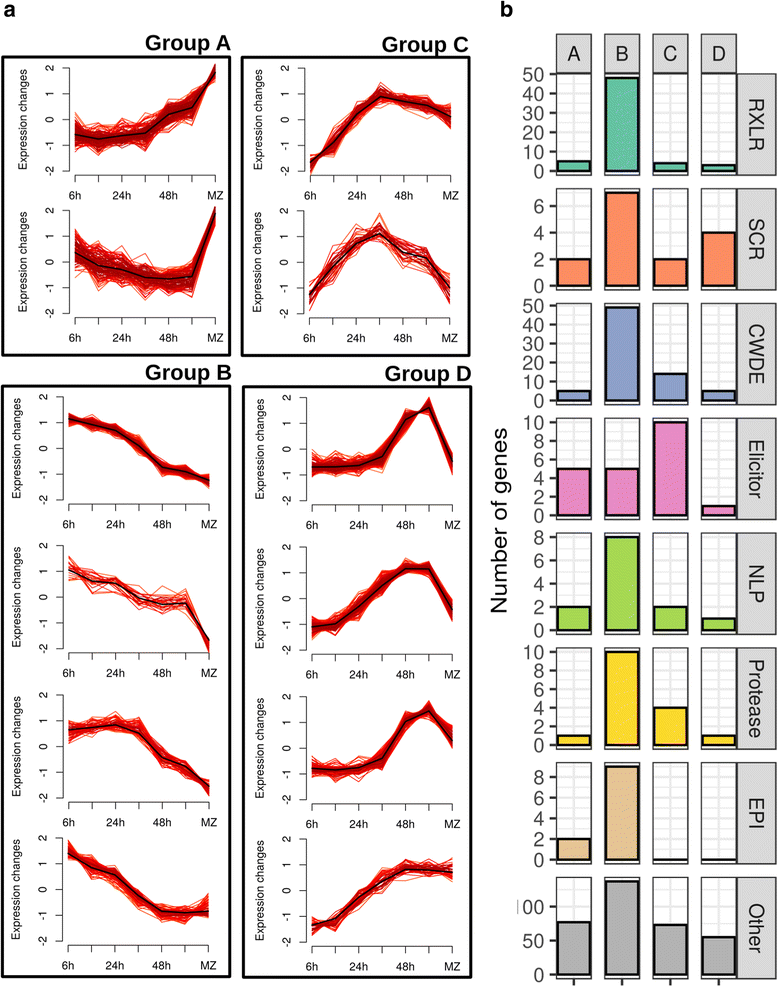
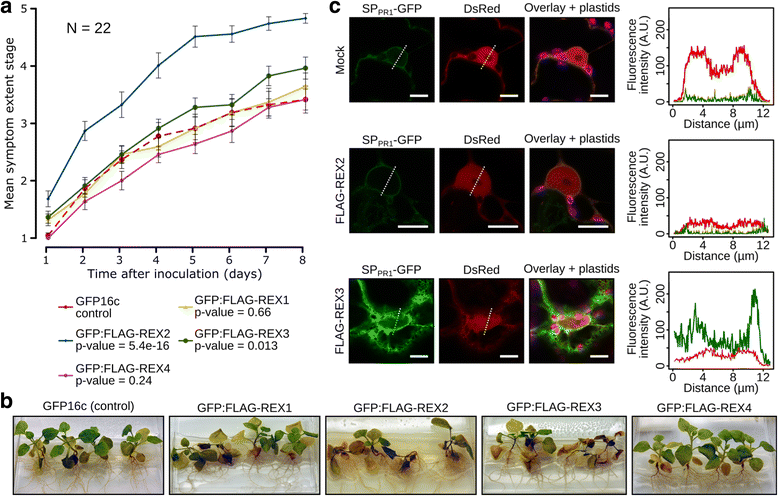
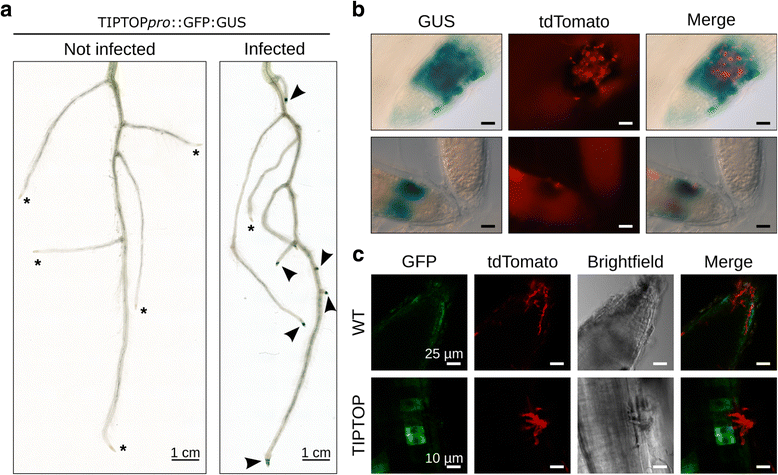
Results: We employed the model plant Nicotiana benthamiana to study the P. palmivora root infections at the cellular and molecular levels. Time-resolved dual transcriptomics revealed different pathogen and host transcriptome dynamics. De novo assembly of P. palmivora transcriptome and semi-automated prediction and annotation of the secretome enabled robust identification of conserved infection-promoting effectors. We show that one of them, REX3, suppresses plant secretion processes. In a survey for early transcriptionally activated plant genes we identified a N. benthamiana gene specifically induced at infected root tips that encodes a peptide with danger-associated molecular features.
Conclusions: These results constitute a major advance in our understanding of P. palmivora diseases and establish extensive resources for P. palmivora pathogenomics, effector-aided resistance breeding and the generation of induced resistance to Phytophthora root infections. Furthermore, our approach to find infection-relevant secreted genes is transferable to other pathogen-host interactions and not restricted to plants.
Keywords: De novo transcriptome assembly; Dual transcriptomics; Effectors; N. benthamiana; Non-model species; P. palmivora; RXLR effectors; Secretome.
Figures



Similar articles
-
Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly related plants.Plant J. 2012 Dec;72(6):882-93. doi: 10.1111/j.1365-313X.2012.05079.x. Epub 2012 Oct 26. Plant J. 2012. PMID: 22709376
-
Differences in transcriptomic responses upon Phytophthora palmivora infection among cultivars reveal potential underlying resistant mechanisms in durian.BMC Plant Biol. 2024 Oct 2;24(1):878. doi: 10.1186/s12870-024-05545-z. BMC Plant Biol. 2024. PMID: 39358741 Free PMC article.
-
LYS12 LysM receptor decelerates Phytophthora palmivora disease progression in Lotus japonicus.Plant J. 2018 Jan;93(2):297-310. doi: 10.1111/tpj.13785. Epub 2017 Dec 22. Plant J. 2018. PMID: 29171909
-
Bud Rot Caused by Phytophthora palmivora: A Destructive Emerging Disease of Oil Palm.Phytopathology. 2016 Apr;106(4):320-9. doi: 10.1094/PHYTO-09-15-0243-RVW. Epub 2016 Mar 16. Phytopathology. 2016. PMID: 26714102 Review.
-
Phytophthora sojae effectors orchestrate warfare with host immunity.Curr Opin Microbiol. 2018 Dec;46:7-13. doi: 10.1016/j.mib.2018.01.008. Epub 2018 Feb 22. Curr Opin Microbiol. 2018. PMID: 29454192 Review.
Cited by
-
Comparative analyses of saprotrophy in Salisapilia sapeloensis and diverse plant pathogenic oomycetes reveal lifestyle-specific gene expression.FEMS Microbiol Ecol. 2020 Oct 24;96(11):fiaa184. doi: 10.1093/femsec/fiaa184. FEMS Microbiol Ecol. 2020. PMID: 32918444 Free PMC article.
-
RPG interacts with E3-ligase CERBERUS to mediate rhizobial infection in Lotus japonicus.PLoS Genet. 2023 Feb 3;19(2):e1010621. doi: 10.1371/journal.pgen.1010621. eCollection 2023 Feb. PLoS Genet. 2023. PMID: 36735729 Free PMC article.
-
Hydrodynamic Shape Changes Underpin Nuclear Rerouting in Branched Hyphae of an Oomycete Pathogen.mBio. 2019 Oct 1;10(5):e01516-19. doi: 10.1128/mBio.01516-19. mBio. 2019. PMID: 31575765 Free PMC article.
-
Chromatin Dynamics Contribute to the Spatiotemporal Expression Pattern of Virulence Genes in a Fungal Plant Pathogen.mBio. 2020 Oct 6;11(5):e02343-20. doi: 10.1128/mBio.02343-20. mBio. 2020. PMID: 33024042 Free PMC article.
-
Haustorium formation and a distinct biotrophic transcriptome characterize infection of Nicotiana benthamiana by the tree pathogen Phytophthora kernoviae.Mol Plant Pathol. 2021 Aug;22(8):954-968. doi: 10.1111/mpp.13072. Epub 2021 May 20. Mol Plant Pathol. 2021. PMID: 34018655 Free PMC article.
References
-
- Erwin DD, Ribeiro OK. Phytophthora diseases worldwide. St. Paul: American Phytopathological Society (APS Press); 1996.
-
- McHau GRA, Coffey MD. Isozyme diversity in Phytophthora palmivora: evidence for a southeast Asian centre of origin. Mycol Res. 1994;98:1035–43. doi: 10.1016/S0953-7562(09)80430-9. - DOI
-
- Scott P, Burgess T, Hardy G. Globalization and Phytophthora. In: Lamour K, editor. Phytophthora: a global perspective. Wallingford: CABI; 2013. pp. 226–32.
-
- Drenth A, Sendall B. Economic impact of Phytophthora diseases in Southeast Asia. In: Drenth A, Guest DI, editors. Diversity and management of Phytophthora in Southeast Asia. Canberra: Australian Centre for International Agricultural Research (ACIAR); 2004. pp. 10–28.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

