Efficient Recreation of t(11;22) EWSR1-FLI1+ in Human Stem Cells Using CRISPR/Cas9
- PMID: 28494941
- PMCID: PMC5425785
- DOI: 10.1016/j.stemcr.2017.04.014
Efficient Recreation of t(11;22) EWSR1-FLI1+ in Human Stem Cells Using CRISPR/Cas9
Abstract
Efficient methodologies for recreating cancer-associated chromosome translocations are in high demand as tools for investigating how such events initiate cancer. The CRISPR/Cas9 system has been used to reconstruct the genetics of these complex rearrangements at native loci while maintaining the architecture and regulatory elements. However, the CRISPR system remains inefficient in human stem cells. Here, we compared three strategies aimed at enhancing the efficiency of the CRISPR-mediated t(11;22) translocation in human stem cells, including mesenchymal and induced pluripotent stem cells: (1) using end-joining DNA processing factors involved in repair mechanisms, or (2) ssODNs to guide the ligation of the double-strand break ends generated by CRISPR/Cas9; and (3) all-in-one plasmid or ribonucleoprotein complex-based approaches. We report that the generation of targeted t(11;22) is significantly increased by using a combination of ribonucleoprotein complexes and ssODNs. The CRISPR/Cas9-mediated generation of targeted t(11;22) in human stem cells opens up new avenues in modeling Ewing sarcoma.
Keywords: CRISPR; Cas9; Ewing sarcoma; MSC; cancer modeling; cancer translocation; disease model; genome engineering; human stem cells; iPSC.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
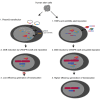
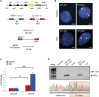
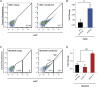


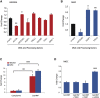
References
-
- Audebert M., Salles B., Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 2004;279:55117–55126. - PubMed
-
- Audebert M., Salles B., Weinfeld M., Calsou P. Involvement of polynucleotide kinase in a poly (ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J. Mol. Biol. 2006;356:257–265. - PubMed
-
- Blasco R.B., Karaca E., Ambrogio C., Cheong T.-C., Karayol E., Minero V.G., Voena C., Chiarle R. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014;9:1219–1227. - PubMed
-
- Bueno C., Montes R., Catalina P., Rodríguez R., Menendez P. Insights into the cellular origin and etiology of the infant pro-B acute lymphoblastic leukemia with MLL-AF4 rearrangement. Leukemia. 2011;25:400–410. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

