Mechanotransduction Dynamics at the Cell-Matrix Interface
- PMID: 28494966
- PMCID: PMC5425358
- DOI: 10.1016/j.bpj.2017.02.027
Mechanotransduction Dynamics at the Cell-Matrix Interface
Abstract
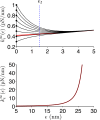
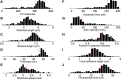
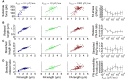
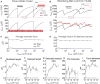
The ability of cells to sense and respond to mechanical cues from the surrounding environment has been implicated as a key regulator of cell differentiation, migration, and proliferation. The extracellular matrix (ECM) is an oft-overlooked component of the interface between cells and their surroundings. Cells assemble soluble ECM proteins into insoluble fibrils with unique mechanical properties that can alter the mechanical cues a cell receives. In this study, we construct a model that predicts the dynamics of cellular traction force generation and subsequent assembly of fibrils of the ECM protein fibronectin (FN). FN fibrils are the primary component in primordial ECM and, as such, FN assembly is a critical component in the cellular mechanical response. The model consists of a network of Hookean springs, each representing an extensible domain within an assembling FN fibril. As actomyosin forces stretch the spring network, simulations predict the resulting traction force and FN fibril formation. The model accurately predicts FN fibril morphometry and demonstrates a mechanism by which FN fibril assembly regulates traction force dynamics in response to mechanical stimuli and varying surrounding substrate stiffness.
Copyright © 2017 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Tug of War at the Cell-Matrix Interface.Biophys J. 2017 May 9;112(9):1739-1741. doi: 10.1016/j.bpj.2017.03.032. Biophys J. 2017. PMID: 28494945 Free PMC article. No abstract available.
References
-
- Chen C.S. Mechanotransduction—a field pulling together? J. Cell Sci. 2008;121:3285–3292. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

