PHD3 is a transcriptional coactivator of HIF-1α in nucleus pulposus cells independent of the PKM2-JMJD5 axis
- PMID: 28495754
- PMCID: PMC5572688
- DOI: 10.1096/fj.201601291R
PHD3 is a transcriptional coactivator of HIF-1α in nucleus pulposus cells independent of the PKM2-JMJD5 axis
Abstract
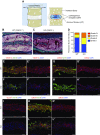
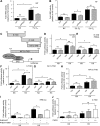
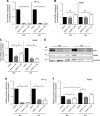
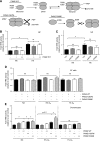
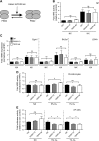
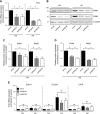
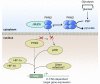
The role of prolyl hydroxylase (PHD)-3 as a hypoxia inducible factor (HIF)-1α cofactor is controversial and remains unknown in skeletal tissues. We investigated whether PHD3 controls HIF-1 transcriptional activity in nucleus pulposus (NP) cells through the pyruvate kinase muscle (PKM)-2-Jumonji domain--containing protein (JMJD5) axis. PHD3-/- mice (12.5 mo old) showed increased incidence of intervertebral disc degeneration with a concomitant decrease in expression of the HIF-1α targets VEGF-A, glucose transporter-1, and lactate dehydrogenase A. PHD3 silencing decreased hypoxic activation of HIF-1α C-terminal transactivation domain (C-TAD), but not HIF-1α-N-terminal-(N)-TAD or HIF-2α-TAD. Moreover, PHD3 suppression in NP cells resulted in decreased HIF-1α enrichment on target promoters and lower expression of select HIF-1 targets. Contrary to other cell types, manipulation of PKM2 and JMJD5 levels had no effect on HIF-1 activity in NP cells. Likewise, stabilization of tetrameric PKM2 by a chemical approach had no effect on PHD3-dependent HIF-1 activity. Coimmunoprecipitation assays showed lack of association between HIF-1α and PKM2 in NP cells. Results support the role of the PHD3 as a cofactor for HIF-1, independent of PKM2-JMJD5.-Schoepflin, Z. R., Silagi, E. S., Shapiro, I. M., Risbud, M. V. PHD3 is a transcriptional coactivator of HIF-1α in nucleus pulposus cells independent of the PKM2-JMJD5 axis.
Keywords: hypoxia; intervertebral disc; skeletal tissue; transcription factor.
© FASEB.
Figures


References
-
- Scaal M. (2016) Early development of the vertebral column. Semin. Cell Dev. Biol. 49, 83–91 - PubMed
-
- Agrawal A., Guttapalli A., Narayan S., Albert T. J., Shapiro I. M., Risbud M. V. (2007) Normoxic stabilization of HIF-1alpha drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am. J. Physiol. Cell Physiol. 293, C621–C631 - PubMed
-
- Risbud M. V., Guttapalli A., Stokes D. G., Hawkins D., Danielson K. G., Schaer T. P., Albert T. J., Shapiro I. M. (2006) Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J. Cell. Biochem. 98, 152–159 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

