Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment
- PMID: 28495837
- PMCID: PMC5461014
- DOI: 10.1083/jcb.201607096
Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment
Abstract
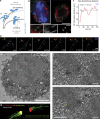
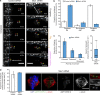
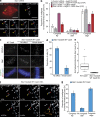
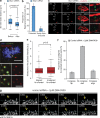
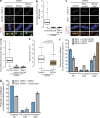
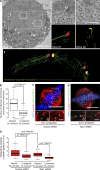
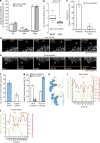
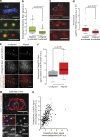
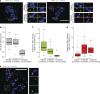
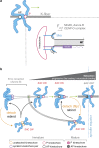
Kinetochores mediate chromosome congression by either sliding along the lattice of spindle microtubules or forming end-on attachments to their depolymerizing plus-ends. By following the fates of individual kinetochores as they congress in live cells, we reveal that the Ska complex is required for a distinct substep of the depolymerization-coupled pulling mechanism. Ska depletion increases the frequency of naturally occurring, force-dependent P kinetochore detachment events, while being dispensable for the initial biorientation and movement of chromosomes. In unperturbed cells, these release events are followed by reattachment and successful congression, whereas in Ska-depleted cells, detached kinetochores remain in a futile reattachment/detachment cycle that prevents congression. We further find that Ska is progressively loaded onto bioriented kinetochore pairs as they congress. We thus propose a model in which kinetochores mature through Ska complex recruitment and that this is required for improved load-bearing capacity and silencing of the spindle assembly checkpoint.
© 2017 Auckland et al.
Figures
References
-
- Abad M.A., Medina B., Santamaria A., Zou J., Plasberg-Hill C., Madhumalar A., Jayachandran U., Redli P.M., Rappsilber J., Nigg E.A., and Jeyaprakash A.A.. 2014. Structural basis for microtubule recognition by the human kinetochore Ska complex. Nat. Commun. 5:2964 10.1038/ncomms3964 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

