Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy
- PMID: 28495853
- PMCID: PMC5582941
- DOI: 10.1152/ajplung.00035.2017
Transgenerational transmission of asthma risk after exposure to environmental particles during pregnancy
Abstract
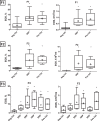
Exposure to environmental particles during pregnancy increases asthma susceptibility of the offspring. We tested the hypothesis that this transmission continues to F2 and F3 generations and occurs via epigenetic mechanisms. We compared allergic susceptibility of three generations of BALB/c offspring after a single maternal exposure during pregnancy to diesel exhaust particles or concentrated urban air particles. After pregnant dams received intranasal instillations of particle suspensions or control, their F1, F2, and F3 offspring were tested in a low-dose ovalbumin protocol for sensitivity to allergic asthma. We found that the elevated susceptibility after maternal exposure to particles during pregnancy persists into F2 and, with lesser magnitude, into F3 generations. This was evident from elevated eosinophil counts in bronchoalveolar lavage (BAL) fluid, histopathological changes of allergic airway disease, and increased BAL levels of IL-5 and IL-13. We have previously shown that dendritic cells (DCs) can mediate transmission of risk upon adoptive transfer. Therefore, we used an enhanced reduced representation bisulfite sequencing protocol to quantify DNA methylation in DCs from each generation. Distinct methylation changes were identified in F1, F2, and F3 DCs. The subset of altered loci shared across the three generations were not linked to known allergy genes or pathways but included a number of genes linked to chromatin modification, suggesting potential interaction with other epigenetic mechanisms (e.g., histone modifications). The data indicate that pregnancy airway exposure to diesel exhaust particles (DEP) triggers a transgenerationally transmitted asthma susceptibility and suggests a mechanistic role for epigenetic alterations in DCs in this process.
Keywords: DNA methylation; asthma; epigenetics; transgenerational.
Copyright © 2017 the American Physiological Society.
Figures

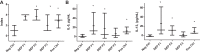





Similar articles
-
Pulmonary exposure to particles during pregnancy causes increased neonatal asthma susceptibility.Am J Respir Cell Mol Biol. 2008 Jan;38(1):57-67. doi: 10.1165/rcmb.2007-0124OC. Epub 2007 Jul 26. Am J Respir Cell Mol Biol. 2008. PMID: 17656681 Free PMC article.
-
Inhalation of diesel exhaust enhances allergen-related eosinophil recruitment and airway hyperresponsiveness in mice.Toxicol Appl Pharmacol. 1998 Jun;150(2):328-37. doi: 10.1006/taap.1998.8437. Toxicol Appl Pharmacol. 1998. PMID: 9653064
-
A mouse model links asthma susceptibility to prenatal exposure to diesel exhaust.J Allergy Clin Immunol. 2014 Jul;134(1):63-72. doi: 10.1016/j.jaci.2013.10.047. Epub 2013 Dec 22. J Allergy Clin Immunol. 2014. PMID: 24365139 Free PMC article.
-
Diesel exhaust particles and airway inflammation.Curr Opin Pulm Med. 2012 Mar;18(2):144-50. doi: 10.1097/MCP.0b013e32834f0e2a. Curr Opin Pulm Med. 2012. PMID: 22234273 Review.
-
Immunotoxicologic analysis of maternal transmission of asthma risk.J Immunotoxicol. 2008 Oct;5(4):445-52. doi: 10.1080/15476910802481765. J Immunotoxicol. 2008. PMID: 19404877 Review.
Cited by
-
Gestational exposure to titanium dioxide, diesel exhaust, and concentrated urban air particles affects levels of specialized pro-resolving mediators in response to allergen in asthma-susceptible neonate lungs.J Toxicol Environ Health A. 2022 Mar 19;85(6):243-261. doi: 10.1080/15287394.2021.2000906. Epub 2021 Nov 21. J Toxicol Environ Health A. 2022. PMID: 34802391 Free PMC article.
-
Asthma and the Missing Heritability Problem: Necessity for Multiomics Approaches in Determining Accurate Risk Profiles.Front Immunol. 2022 May 25;13:822324. doi: 10.3389/fimmu.2022.822324. eCollection 2022. Front Immunol. 2022. PMID: 35693821 Free PMC article. Review.
-
Transcriptomic and epigenomic effects of insoluble particles on J774 macrophages.Epigenetics. 2021 Oct;16(10):1053-1070. doi: 10.1080/15592294.2020.1834925. Epub 2020 Oct 30. Epigenetics. 2021. PMID: 33054565 Free PMC article.
-
Fetotoxicity of Nanoparticles: Causes and Mechanisms.Nanomaterials (Basel). 2021 Mar 19;11(3):791. doi: 10.3390/nano11030791. Nanomaterials (Basel). 2021. PMID: 33808794 Free PMC article. Review.
-
Grandmaternal allergen sensitization reprograms epigenetic and airway responses to allergen in second-generation offspring.Am J Physiol Lung Cell Mol Physiol. 2023 Dec 1;325(6):L776-L787. doi: 10.1152/ajplung.00103.2023. Epub 2023 Oct 10. Am J Physiol Lung Cell Mol Physiol. 2023. PMID: 37814791 Free PMC article.
References
-
- Brandsma C-A, van den Berge M, Postma DS, Jonker MR, Brouwer S, Paré PD, Sin DD, Bossé Y, Laviolette M, Karjalainen J, Fehrmann RSN, Nickle DC, Hao K, Spanjer AIR, Timens W, Franke L. A large lung gene expression study identifying fibulin-5 as a novel player in tissue repair in COPD. Thorax 70: 21–32, 2015. doi:10.1136/thoraxjnl-2014-205091. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

