Dynamic expression and regulatory mechanism of TGF-β signaling in chicken embryonic stem cells differentiating into spermatogonial stem cells
- PMID: 28495881
- PMCID: PMC6434085
- DOI: 10.1042/BSR20170179
Dynamic expression and regulatory mechanism of TGF-β signaling in chicken embryonic stem cells differentiating into spermatogonial stem cells
Abstract
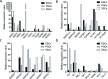
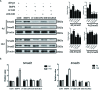
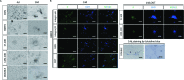
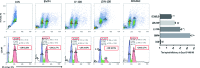
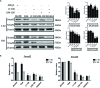
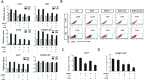
The present study investigated the dynamic expression and regulatory mechanism of transforming growth factor β (TGF-β) signaling involved in embryonic stem cells (ESCs) differentiation into male germ cells. Candidate genes involved in TGF-β signaling pathway were screened from RNA-sequencing (RNA-seq), which were further validated by quantitative real-time PCR (qRT-PCR). Bone morphogenetic protein 4 (BMP4) was used to induce differentiation of ESCs in vitro Inhibition of TGF-β signaling pathway was reflected by Western blot of SMAD2 and SMAD5 expression. Differentiating efficiency of germ cells was evaluated by immunofluorescence and fluorescence-activated cell sorting (FACS). Germ cell marker genes were assessed by qRT-PCR in the differentiation process, with activation or inhibition of TGF-β signaling pathway. In the process of in vitro induction, SMAD2 and SMAD5 were found to significantly up-regulated in BMP4 group versus the control and inhibition groups after 4 and 14 days. Expression of CKIT, CVH, DAZL, STRA8, and INTEGRIN α6 were significantly increased in the BMP4 group compared with the control group, while down-regulated in the inhibition groups. The proportion of germ cell-like cells was decreased from 17.9% to 2.2% after 4 days induction, and further decreased from 14.1% to 2.1% after 14 days induction. Correspondingly, expression of marker genes in germ cells was significantly lower. In vivo inhibition of TGF-β signaling pathway reduced germ cells formation from 5.5% to 1.6%, and down-regulated the expression of CKIT, CVH, DAZL, STRA8, and INTEGRIN α6 In conclusion, our study reveals the mechanism regulating spermatogonial stem cells (SSCs) and lays the basis for further understanding of the regulatory network.
Keywords: Embryonic stem cells; Primordial germ cells; RNA-seq; Spermatogonial stem cells; TGF-β signaling.
© 2017 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


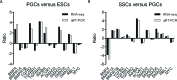

Similar articles
-
Effects of the Transforming Growth Factor Beta Signaling Pathway on the Differentiation of Chicken Embryonic Stem Cells into Male Germ Cells.Cell Reprogram. 2016 Nov;18(6):401-410. doi: 10.1089/cell.2016.0019. Cell Reprogram. 2016. PMID: 27906584 Free PMC article.
-
Wnt signaling pathway regulates differentiation of chicken embryonic stem cells into spermatogonial stem cells via Wnt5a.J Cell Biochem. 2018 Feb;119(2):1689-1701. doi: 10.1002/jcb.26329. Epub 2017 Sep 25. J Cell Biochem. 2018. PMID: 28786525
-
Distinct roles of retinoic acid and BMP4 pathways in the formation of chicken primordial germ cells and spermatogonial stem cells.Food Funct. 2019 Nov 1;10(11):7152-7163. doi: 10.1039/c9fo01485c. Epub 2019 Oct 9. Food Funct. 2019. PMID: 31596288
-
Differential response of epiblast stem cells to Nodal and Activin signalling: a paradigm of early endoderm development in the embryo.Philos Trans R Soc Lond B Biol Sci. 2014 Dec 5;369(1657):20130550. doi: 10.1098/rstb.2013.0550. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25349457 Free PMC article. Review.
-
TGF-β Family Signaling in Embryonic and Somatic Stem-Cell Renewal and Differentiation.Cold Spring Harb Perspect Biol. 2017 Jul 5;9(7):a022186. doi: 10.1101/cshperspect.a022186. Cold Spring Harb Perspect Biol. 2017. PMID: 28108485 Free PMC article. Review.
Cited by
-
Strand-Specific RNA Sequencing Reveals Gene Expression Patterns in F1 Chick Breast Muscle and Liver after Hatching.Animals (Basel). 2024 Apr 29;14(9):1335. doi: 10.3390/ani14091335. Animals (Basel). 2024. PMID: 38731340 Free PMC article.
-
Efficient generation of male germ-like cells derived during co-culturing of adipose-derived mesenchymal stem cells with Sertoli cells under retinoic acid and testosterone induction.Stem Cell Res Ther. 2019 Mar 13;10(1):91. doi: 10.1186/s13287-019-1181-5. Stem Cell Res Ther. 2019. PMID: 30867048 Free PMC article.
-
Fate Decisions of Chicken Primordial Germ Cells (PGCs): Development, Integrity, Sex Determination, and Self-Renewal Mechanisms.Genes (Basel). 2023 Feb 28;14(3):612. doi: 10.3390/genes14030612. Genes (Basel). 2023. PMID: 36980885 Free PMC article. Review.
-
Screening of differentially expressed immune-related genes from spleen of broilers fed with probiotic Bacillus cereus PAS38 based on suppression subtractive hybridization.PLoS One. 2019 Dec 23;14(12):e0226829. doi: 10.1371/journal.pone.0226829. eCollection 2019. PLoS One. 2019. PMID: 31869398 Free PMC article.
-
Trajectory modeling of endothelial-to-mesenchymal transition reveals galectin-3 as a mediator in pulmonary fibrosis.Cell Death Dis. 2021 Mar 26;12(4):327. doi: 10.1038/s41419-021-03603-0. Cell Death Dis. 2021. PMID: 33771973 Free PMC article.
References
-
- Yue J., Mulder K.M., Yue J. and Mulder K.M. (2001) Transforming growth factor-β signal transduction in epithelial cells. Pharmacol. Ther. 91, 1–34 - PubMed
-
- Nohe A., Keating E., Knaus P. and Petersen N.O. (2004) Signal transduction of bone morphogenetic protein receptors. Cell. Signal. 16, 291–299 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

