Bacterial Biosensors for in Vivo Spatiotemporal Mapping of Root Secretion
- PMID: 28495892
- PMCID: PMC5490882
- DOI: 10.1104/pp.16.01302
Bacterial Biosensors for in Vivo Spatiotemporal Mapping of Root Secretion
Abstract
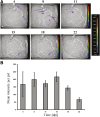
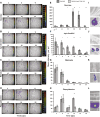
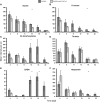
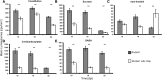
Plants engineer the rhizosphere to their advantage by secreting various nutrients and secondary metabolites. Coupling transcriptomic and metabolomic analyses of the pea (Pisum sativum) rhizosphere, a suite of bioreporters has been developed in Rhizobium leguminosarum bv viciae strain 3841, and these detect metabolites secreted by roots in space and time. Fourteen bacterial lux fusion bioreporters, specific for sugars, polyols, amino acids, organic acids, or flavonoids, have been validated in vitro and in vivo. Using different bacterial mutants (nodC and nifH), the process of colonization and symbiosis has been analyzed, revealing compounds important in the different steps of the rhizobium-legume association. Dicarboxylates and sucrose are the main carbon sources within the nodules; in ineffective (nifH) nodules, particularly low levels of sucrose were observed, suggesting that plant sanctions affect carbon supply to nodules. In contrast, high myo-inositol levels were observed prior to nodule formation and also in nifH senescent nodules. Amino acid biosensors showed different patterns: a γ-aminobutyrate biosensor was active only inside nodules, whereas the phenylalanine bioreporter showed a high signal also in the rhizosphere. The bioreporters were further validated in vetch (Vicia hirsuta), producing similar results. In addition, vetch exhibited a local increase of nod gene-inducing flavonoids at sites where nodules developed subsequently. These bioreporters will be particularly helpful in understanding the dynamics of root exudation and the role of different molecules secreted into the rhizosphere.
© 2017 The author(s). All Rights Reserved.
Figures


References
-
- Badri DV, Weir TL, van der Lelie D, Vivanco JM (2009) Rhizosphere chemical dialogues: plant-microbe interactions. Curr Opin Biotechnol 20: 642–650 - PubMed
-
- Baetz U, Martinoia E (2014) Root exudates: the hidden part of plant defense. Trends Plant Sci 19: 90–98 - PubMed
-
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266 - PubMed
-
- Beringer JE. (1974) R factor transfer in Rhizobium leguminosarum. J Gen Microbiol 84: 188–198 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

