Pias3 is necessary for dorso-ventral patterning and visual response of retinal cones but is not required for rod photoreceptor differentiation
- PMID: 28495965
- PMCID: PMC5483026
- DOI: 10.1242/bio.024679
Pias3 is necessary for dorso-ventral patterning and visual response of retinal cones but is not required for rod photoreceptor differentiation
Abstract
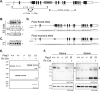
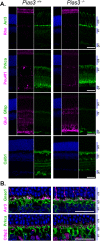
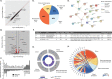
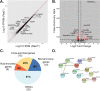
Protein inhibitor of activated Stat 3 (Pias3) is implicated in guiding specification of rod and cone photoreceptors through post-translational modification of key retinal transcription factors. To investigate its role during retinal development, we deleted exon 2-5 of the mouse Pias3 gene, which resulted in complete loss of the Pias3 protein. Pias3-/- mice did not show any overt phenotype, and retinal lamination appeared normal even at 18 months. We detected reduced photopic b-wave amplitude by electroretinography following green light stimulation of postnatal day (P)21 Pias3-/- retina, suggesting a compromised visual response of medium wavelength (M) cones. No change was evident in response of short wavelength (S) cones or rod photoreceptors until 7 months. Increased S-opsin expression in the M-cone dominant dorsal retina suggested altered distribution of cone photoreceptors. Transcriptome profiling of P21 and 18-month-old Pias3-/- retina revealed aberrant expression of a subset of photoreceptor genes. Our studies demonstrate functional redundancy in SUMOylation-associated transcriptional control mechanisms and identify a specific, though limited, role of Pias3 in modulating spatial patterning and optimal function of cone photoreceptor subtypes in the mouse retina.
Keywords: Cell type specification; Gene regulation; Mouse knockout; Retina development; SUMOylation; Vision.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



References
-
- Applebury M. L., Antoch M. P., Baxter L. C., Chun L. L. Y., Falk J. D., Farhangfar F., Kage K., Krzystolik M. G., Lyass L. A. and Robbins J. T. (2000). The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron 27, 513-523. 10.1016/S0896-6273(00)00062-3 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

