High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding
- PMID: 28495970
- PMCID: PMC5448226
- DOI: 10.1073/pnas.1700557114
High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding
Abstract
The bacterial adaptive immune system CRISPR-Cas9 has been appropriated as a versatile tool for editing genomes, controlling gene expression, and visualizing genetic loci. To analyze Cas9's ability to bind DNA rapidly and specifically, we generated multiple libraries of potential binding partners for measuring the kinetics of nuclease-dead Cas9 (dCas9) interactions. Using a massively parallel method to quantify protein-DNA interactions on a high-throughput sequencing flow cell, we comprehensively assess the effects of combinatorial mismatches between guide RNA (gRNA) and target nucleotides, both in the seed and in more distal nucleotides, plus disruption of the protospacer adjacent motif (PAM). We report two consequences of PAM-distal mismatches: reversal of dCas9 binding at long time scales, and synergistic changes in association kinetics when other gRNA-target mismatches are present. Together, these observations support a model for Cas9 specificity wherein gRNA-DNA mismatches at PAM-distal bases modulate different biophysical parameters that determine association and dissociation rates. The methods we present decouple aspects of kinetic and thermodynamic properties of the Cas9-DNA interaction and broaden the toolkit for investigating off-target binding behavior.
Keywords: CRISPR; DNA; kinetics; molecular biophysics; sequencing.
Conflict of interest statement
Conflict of interest statement: S.H.S. is an employee of Caribou Biosciences, Inc. and an inventor on patents and patent applications related to CRISPR-Cas systems and applications thereof. J.A.D. is a cofounder of Editas Medicine, Intellia Therapeutics, and Caribou Biosciences and a scientific advisor to Caribou, Intellia, eFFECTOR Therapeutics, and Driver. J.A.D. receives funding from Roche, Pfizer, the Paul Allen Institute, and the Keck Foundation.
Figures
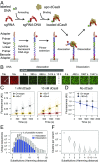
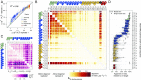
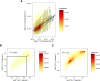

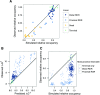







References
-
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. - PubMed
-
- Wang H, La Russa M, Qi LS. CRISPR/Cas9 in genome editing and beyond. Annu Rev Biochem. 2016;85:227–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

