Genetic structure of Cercospora beticola populations on Beta vulgaris in New York and Hawaii
- PMID: 28496148
- PMCID: PMC5431814
- DOI: 10.1038/s41598-017-01929-4
Genetic structure of Cercospora beticola populations on Beta vulgaris in New York and Hawaii
Abstract
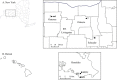
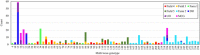
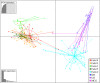
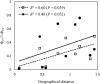
Cercospora leaf spot (CLS), caused by Cercospora beticola, is a major disease of Beta vulgaris worldwide. No sexual stage is known for C. beticola but in its asexual form it overwinters on infected plant debris as pseudostromata, and travels short distances by rain splash-dispersed conidiospores. Cercospora beticola infects a broad range of host species and may be seedborne. The relative contribution of these inoculum sources to CLS epidemics on table beet is not well understood. Pathogen isolates collected from table beet, Swiss chard and common lambsquarters in mixed-cropping farms and monoculture fields in New York and Hawaii, USA, were genotyped (n = 600) using 12 microsatellite markers. All isolates from CLS symptoms on lambsquarters were identified as C. chenopodii. Sympatric populations of C. beticola derived from Swiss chard and table beet were not genetically differentiated. Results suggested that local (within field) inoculum sources may be responsible for the initiation of CLS epidemics in mixed-cropping farms, whereas external sources of inoculum may be contributing to CLS epidemics in the monoculture fields in New York. New multiplex PCR assays were developed for mating-type determination for C. beticola. Implications of these findings for disease management are discussed.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- McDonald BA. How can research on pathogen population biology suggest disease management strategies? The example of barley scald (Rhynchosporium commune) Plant Pathol. 2015;64:1005–1013. doi: 10.1111/ppa.12415. - DOI
-
- Linde C. Population genetic analyses of plant pathogens: new challenges and opportunities. Australas. Plant Pathol. 2010;39:23–28. doi: 10.1071/AP09061. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

