Long non-coding RNA PARTICLE bridges histone and DNA methylation
- PMID: 28496150
- PMCID: PMC5431818
- DOI: 10.1038/s41598-017-01875-1
Long non-coding RNA PARTICLE bridges histone and DNA methylation
Abstract
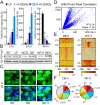
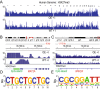
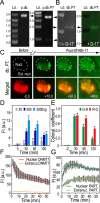
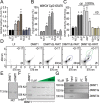
PARTICLE (Gene PARTICL- 'Promoter of MAT2A-Antisense RadiaTion Induced Circulating LncRNA) expression is transiently elevated following low dose irradiation typically encountered in the workplace and from natural sources. This long non-coding RNA recruits epigenetic silencers for cis-acting repression of its neighbouring Methionine adenosyltransferase 2A gene. It now emerges that PARTICLE operates as a trans-acting mediator of DNA and histone lysine methylation. Chromatin immunoprecipitation sequencing (ChIP-seq) and immunological evidence established elevated PARTICLE expression linked to increased histone 3 lysine 27 trimethylation. Live-imaging of dbroccoli-PARTICLE revealing its dynamic association with DNA methyltransferase 1 was confirmed by flow cytometry, immunoprecipitation and direct competitive binding interaction through electrophoretic mobility shift assay. Acting as a regulatory docking platform, the long non-coding RNA PARTICLE serves to interlink epigenetic modification machineries and represents a compelling innovative component necessary for gene silencing on a global scale.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
PARTICLE triplexes cluster in the tumor suppressor WWOX and may extend throughout the human genome.Sci Rep. 2017 Aug 2;7(1):7163. doi: 10.1038/s41598-017-07295-5. Sci Rep. 2017. PMID: 28769061 Free PMC article.
-
PARTICLE - The RNA podium for genomic silencers.J Cell Physiol. 2019 Nov;234(11):19464-19470. doi: 10.1002/jcp.28739. Epub 2019 May 6. J Cell Physiol. 2019. PMID: 31058319
-
PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation.Cell Rep. 2015 Apr 21;11(3):474-85. doi: 10.1016/j.celrep.2015.03.043. Cell Rep. 2015. PMID: 25900080
-
Epigenetic regulation in human melanoma: past and future.Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
Cited by
-
LncRNA MIR17HG Suppresses Breast Cancer Proliferation and Migration as ceRNA to Target FAM135A by Sponging miR-454-3p.Mol Biotechnol. 2023 Dec;65(12):2071-2085. doi: 10.1007/s12033-023-00706-1. Epub 2023 Mar 21. Mol Biotechnol. 2023. PMID: 36943627 Free PMC article.
-
Chromatin modifications in metabolic disease: Potential mediators of long-term disease risk.Wiley Interdiscip Rev Syst Biol Med. 2018 Jul;10(4):e1416. doi: 10.1002/wsbm.1416. Epub 2018 Jan 25. Wiley Interdiscip Rev Syst Biol Med. 2018. PMID: 29369528 Free PMC article. Review.
-
Investigating the role of long non-coding RNA in hypertrophic cardiomyopathy.bioRxiv [Preprint]. 2025 Jul 31:2025.07.26.666851. doi: 10.1101/2025.07.26.666851. bioRxiv. 2025. PMID: 40766666 Free PMC article. Preprint.
-
Methodological Challenges in Developmental Human Behavioral Epigenetics: Insights Into Study Design.Front Behav Neurosci. 2018 Nov 23;12:286. doi: 10.3389/fnbeh.2018.00286. eCollection 2018. Front Behav Neurosci. 2018. PMID: 30532698 Free PMC article.
-
LINC01137/miR-186-5p/WWOX: a novel axis identified from WWOX-related RNA interactome in bladder cancer.Front Genet. 2023 Jul 13;14:1214968. doi: 10.3389/fgene.2023.1214968. eCollection 2023. Front Genet. 2023. PMID: 37519886 Free PMC article.
References
-
- Alvarez L, Sanchez-Gongora E, Mingorance J, Pajares MA, Mato JM. Characterization of rat liver-specific methionine adenosyltransferase gene promoter. Role of distal upstream cis-acting elements in the regulation of the transcriptional activity. J Biol Chem. 1997;272:22875–22883. doi: 10.1074/jbc.272.36.22875. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

