A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source
- PMID: 28496395
- PMCID: PMC5421409
- DOI: 10.1007/s11306-016-1157-8
A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source
Abstract
Introduction: Advances in high-resolution mass spectrometry have created renewed interest for studying global lipid biochemistry in disease and biological systems.
Objectives: Here, we present an untargeted 30 min. LC-MS/MS platform that utilizes positive/negative polarity switching to perform unbiased data dependent acquisitions (DDA) via higher energy collisional dissociation (HCD) fragmentation to profile more than 1000-1500 lipid ions mainly from methyl-tert-butyl ether (MTBE) or chloroform:methanol extractions.
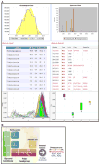
Methods: The platform uses C18 reversed-phase chromatography coupled to a hybrid QExactive Plus/HF Orbitrap mass spectrometer and the entire procedure takes ~10 h from lipid extraction to identification/quantification for a data set containing 12 samples (~4 h for a single sample). Lipids are identified by both accurate precursor ion mass and fragmentation features and quantified using Lipid-Search and Elements software.
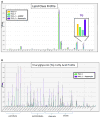
Results: Using this approach, we are able to profile intact lipid ions from up to 18 different main lipid classes and 66 subclasses. We show several studies from different biological sources, including cultured cancer cells, resected tissues from mice such as lung and breast tumors and biological fluids such as plasma and urine.
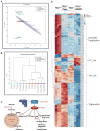
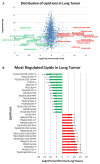
Conclusions: Using mouse embryonic fibroblasts, we showed that TSC2-/- KD significantly abrogates lipid biosynthesis and that rapamycin can rescue triglyceride (TG) lipids and we show that SREBP-/- shuts down lipid biosynthesis significantly via mTORC1 signaling pathways. We show that in mouse EGFR driven lung tumors, a large number of TGs and phosphatidylmethanol (PMe) lipids are elevated while some phospholipids (PLs) show some of the largest decrease in lipid levels from ~ 2000 identified lipid ions. In addition, we identified more than 1500 unique lipid species from human blood plasma.
Keywords: Biomarkers; Cancer; Disease; LC-MS/MS; Lipidomics; Mass spectrometry; Polarity switching; Profiling; Quantification; Shotgun.
Figures


References
-
- AHMED Z, MAYR M, ZEESHAN S, DANDEKAR T, MUEL-LER MJ, FEKETE A. Lipid-Pro: A computational lipid identification solution for untargeted lipidomics on data-independent acquisition tandem mass spectrometry platforms. Bioinformatics (Oxford, England) 2015;31:1150–1153. - PubMed
-
- Asara JM, Xu Y, Breitkopf SB, Yuan M, Ricoult SJ, Manning BD. Preparing Biological Samples for Metabolomics and Lipidomics, Can We Start with Just One Sample? Association of Biomolecular Resource Facilities; Ft. Lauderdale: 2016.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
