Systematic analysis of the lysine acetylome reveals diverse functions of lysine acetylation in the oleaginous yeast Yarrowia lipolytica
- PMID: 28497289
- PMCID: PMC5427063
- DOI: 10.1186/s13568-017-0393-2
Systematic analysis of the lysine acetylome reveals diverse functions of lysine acetylation in the oleaginous yeast Yarrowia lipolytica
Abstract
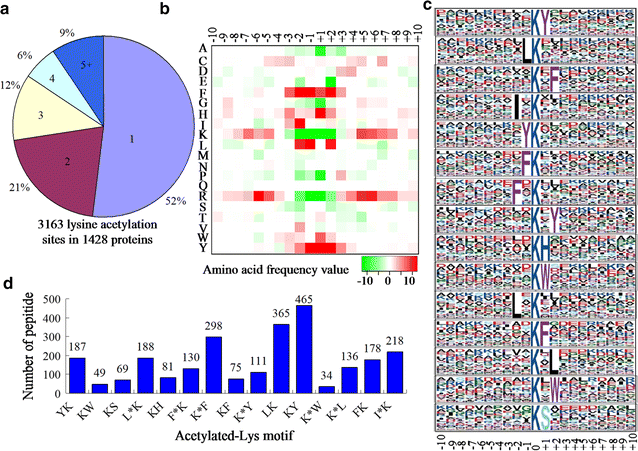
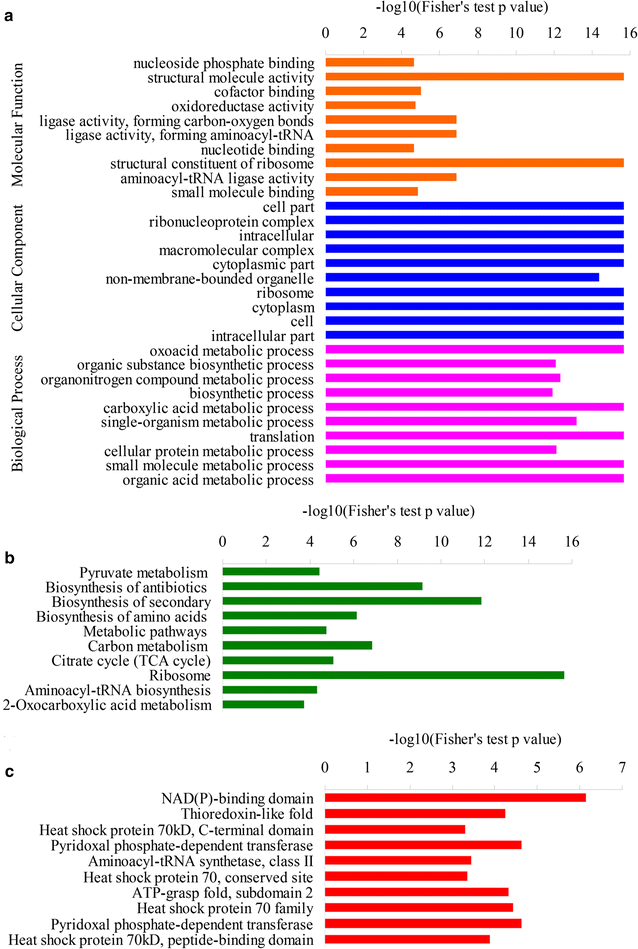
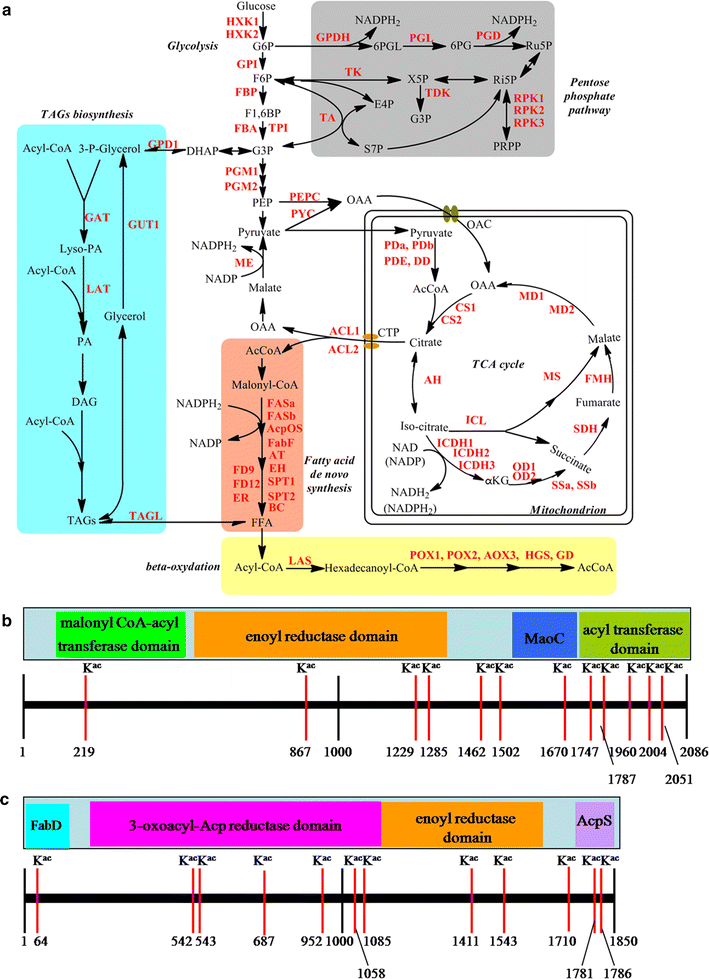
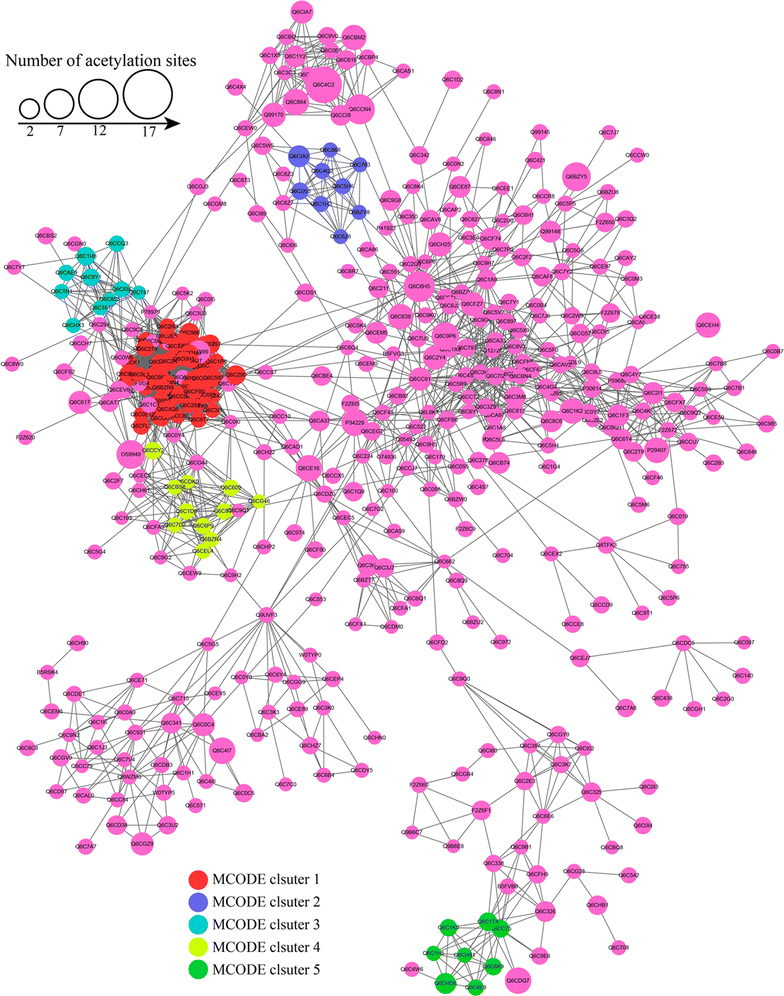
Lysine acetylation of proteins, a major post-translational modification, plays a critical regulatory role in almost every aspects in both eukaryotes and prokaryotes. Yarrowia lipolytica, an oleaginous yeast, is considered as a model for bio-oil production due to its ability to accumulate a large amount of lipids. However, the function of lysine acetylation in this organism is elusive. Here, we performed a global acetylproteome analysis of Y. lipolytica ACA-DC 50109. In total, 3163 lysine acetylation sites were identified in 1428 proteins, which account for 22.1% of the total proteins in the cell. Fifteen conserved acetylation motifs were detected. The acetylated proteins participate in a wide variety of biological processes. Notably, a total of 65 enzymes involved in lipid biosynthesis were found to be acetylated. The acetylation sites are distributed in almost every type of conserved domains in the multi-enzymatic complexes of fatty acid synthetases. The provided dataset probably illuminates the crucial role of reversible acetylation in oleaginous microorganisms, and serves as an important resource for exploring the physiological role of lysine acetylation in eukaryotes.
Keywords: Acetylproteome; Lipid biosynthesis; Lysine acetylation; Oleaginous yeast; Yarrowia lipolytica.
Figures






References
-
- Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinform Chap. 2011;13:15–24. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

