Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain
- PMID: 28497798
- PMCID: PMC5437310
- DOI: 10.1038/ncomms15233
Phosphorylation induces sequence-specific conformational switches in the RNA polymerase II C-terminal domain
Abstract
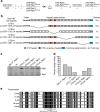
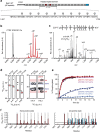
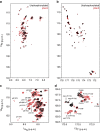
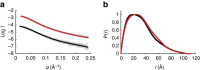
The carboxy-terminal domain (CTD) of the RNA polymerase II (Pol II) large subunit cycles through phosphorylation states that correlate with progression through the transcription cycle and regulate nascent mRNA processing. Structural analyses of yeast and mammalian CTD are hampered by their repetitive sequences. Here we identify a region of the Drosophila melanogaster CTD that is essential for Pol II function in vivo and capitalize on natural sequence variations within it to facilitate structural analysis. Mass spectrometry and NMR spectroscopy reveal that hyper-Ser5 phosphorylation transforms the local structure of this region via proline isomerization. The sequence context of this switch tunes the activity of the phosphatase Ssu72, leading to the preferential de-phosphorylation of specific heptads. Together, context-dependent conformational switches and biased dephosphorylation suggest a mechanism for the selective recruitment of cis-proline-specific regulatory factors and region-specific modulation of the CTD code that may augment gene regulation in developmentally complex organisms.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

