Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome
- PMID: 28498102
- PMCID: PMC5462543
- DOI: 10.7554/eLife.23717
Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome
Abstract
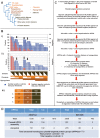
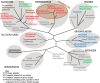
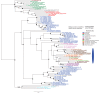
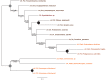
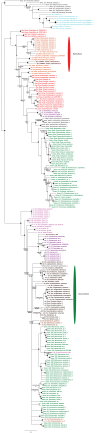
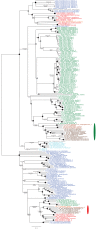
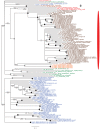
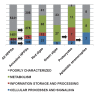
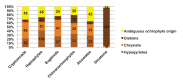
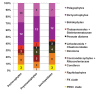
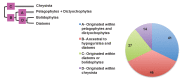
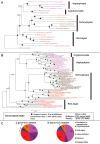
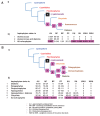
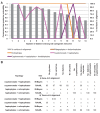
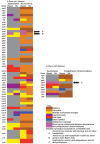
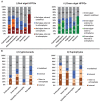
Plastids are supported by a wide range of proteins encoded within the nucleus and imported from the cytoplasm. These plastid-targeted proteins may originate from the endosymbiont, the host, or other sources entirely. Here, we identify and characterise 770 plastid-targeted proteins that are conserved across the ochrophytes, a major group of algae including diatoms, pelagophytes and kelps, that possess plastids derived from red algae. We show that the ancestral ochrophyte plastid proteome was an evolutionary chimera, with 25% of its phylogenetically tractable nucleus-encoded proteins deriving from green algae. We additionally show that functional mixing of host and plastid proteomes, such as through dual-targeting, is an ancestral feature of plastid evolution. Finally, we detect a clear phylogenetic signal from one ochrophyte subgroup, the lineage containing pelagophytes and dictyochophytes, in plastid-targeted proteins from another major algal lineage, the haptophytes. This may represent a possible serial endosymbiosis event deep in eukaryotic evolutionary history.
Keywords: HPPG; algae; cell biology; chromalveolate; diatom; evolutionary biology; genomics; none; plastid protein import; shopping bag model.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures























References
-
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research. 2016;44:W3–W10. doi: 10.1093/nar/gkw343. - DOI - PMC - PubMed
-
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou S, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, Medina M, Montsant A, Obornik M, Parker MS, Palenik B, Pazour GJ, Richardson PM, Rynearson TA, Saito MA, Schwartz DC, Thamatrakoln K, Valentin K, Vardi A, Wilkerson FP, Rokhsar DS. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

