Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis
- PMID: 28498104
- PMCID: PMC5519331
- DOI: 10.7554/eLife.23471
Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis
Abstract
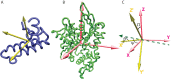
The interaction between the Heat Shock Proteins 70 and 40 is at the core of the ATPase regulation of the chaperone machinery that maintains protein homeostasis. However, the structural details of the interaction remain elusive and contrasting models have been proposed for the transient Hsp70/Hsp40 complexes. Here we combine molecular simulations based on both coarse-grained and atomistic models with coevolutionary sequence analysis to shed light on this problem by focusing on the bacterial DnaK/DnaJ system. The integration of these complementary approaches resulted in a novel structural model that rationalizes previous experimental observations. We identify an evolutionarily conserved interaction surface formed by helix II of the DnaJ J-domain and a structurally contiguous region of DnaK, involving lobe IIA of the nucleotide binding domain, the inter-domain linker, and the β-basket of the substrate binding domain.
Keywords: biophysics; computational biology; heat shock protein; molecular modeling; none; protein coevolution; protein complex; structural biology; systems biology; transient interaction.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures


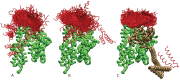
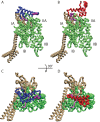





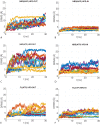
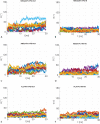




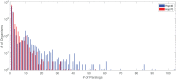
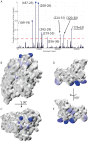





Similar articles
-
Cytosolic and ER J-domains of mammalian and parasitic origin can functionally interact with DnaK.Int J Biochem Cell Biol. 2007;39(4):736-51. doi: 10.1016/j.biocel.2006.11.006. Epub 2006 Nov 23. Int J Biochem Cell Biol. 2007. PMID: 17239655 Free PMC article.
-
A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein.J Biol Chem. 2015 Apr 3;290(14):8849-62. doi: 10.1074/jbc.M114.596288. Epub 2015 Jan 28. J Biol Chem. 2015. PMID: 25635056 Free PMC article.
-
Structure and energetics of an allele-specific genetic interaction between dnaJ and dnaK: correlation of nuclear magnetic resonance chemical shift perturbations in the J-domain of Hsp40/DnaJ with binding affinity for the ATPase domain of Hsp70/DnaK.Biochemistry. 2003 May 6;42(17):4926-36. doi: 10.1021/bi027070y. Biochemistry. 2003. PMID: 12718534
-
The Hsp70 chaperone machines of Escherichia coli: a paradigm for the repartition of chaperone functions.Mol Microbiol. 2007 Nov;66(4):840-57. doi: 10.1111/j.1365-2958.2007.05961.x. Epub 2007 Oct 4. Mol Microbiol. 2007. PMID: 17919282 Review.
-
Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes.Structure. 2016 Jul 6;24(7):1014-30. doi: 10.1016/j.str.2016.05.011. Epub 2016 Jun 23. Structure. 2016. PMID: 27345933 Free PMC article. Review.
Cited by
-
Pseudophosphorylation of single residues of the J-domain of DNAJA2 regulates the holding/folding balance of the Hsc70 system.Protein Sci. 2024 Aug;33(8):e5105. doi: 10.1002/pro.5105. Protein Sci. 2024. PMID: 39012012 Free PMC article.
-
Mitotic phosphorylation regulates Hsp72 spindle localization by uncoupling ATP binding from substrate release.Sci Signal. 2018 Aug 14;11(543):eaao2464. doi: 10.1126/scisignal.aao2464. Sci Signal. 2018. PMID: 30108182 Free PMC article.
-
Coevolutionary Analysis of Protein Subfamilies by Sequence Reweighting.Entropy (Basel). 2020 Jan 23;21(11):1127. doi: 10.3390/e21111127. Epub 2019 Nov 16. Entropy (Basel). 2020. PMID: 32002010 Free PMC article.
-
Engineering repressors with coevolutionary cues facilitates toggle switches with a master reset.Nucleic Acids Res. 2019 Jun 4;47(10):5449-5463. doi: 10.1093/nar/gkz280. Nucleic Acids Res. 2019. PMID: 31162606 Free PMC article.
-
The Plasmodium falciparum exported J domain proteins fine-tune human and malarial Hsp70s: pathological exploitation of proteostasis machinery.Front Mol Biosci. 2023 Jun 30;10:1216192. doi: 10.3389/fmolb.2023.1216192. eCollection 2023. Front Mol Biosci. 2023. PMID: 37457831 Free PMC article. Review.
References
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

