The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus
- PMID: 28498729
- PMCID: PMC5810505
- DOI: 10.1080/21505594.2017.1330240
The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus
Abstract
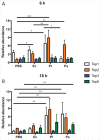
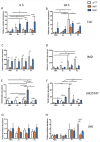
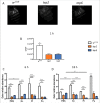
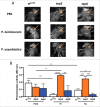
Previous and recent investigations on the innate immune response of Drosophila have identified certain mechanisms that promote pathogen elimination. However, the function of Thioester-containing proteins (TEPs) in the fly still remains elusive. Recently we have shown the contribution of TEP4 in the antibacterial immune defense of Drosophila against non-pathogenic E. coli, and the pathogens Photorhabdus luminescens and P. asymbiotica. Here we studied the function of Tep genes in both humoral and cellular immunity upon E. coli and Photorhabdus infection. We found that while Tep2 is induced after Photorhabdus and E. coli infection; Tep6 is induced by P. asymbiotica only. Moreover, functional ablation of hemocytes results in significantly low transcript levels of Tep2 and Tep6 in response to Photorhabdus. We show that Tep2 and Tep6 loss-of-function mutants have prolonged survival against P. asymbiotica, Tep6 mutants survive better the infection of P. luminescens, and both tep mutants are resistant to E. coli and Photorhabdus. We also find a distinct pattern of immune signaling pathway induction in E. coli or Photorhabdus infected Tep2 and Tep6 mutants. We further show that Tep2 and Tep6 participate in the activation of hemocytes in Drosophila responding to Photorhabdus. Finally, inactivation of Tep2 or Tep6 affects phagocytosis and melanization in flies infected with Photorhabdus. Our results indicate that distinct Tep genes might be involved in different yet crucial functions in the Drosophila antibacterial immune response.
Keywords: Drosophila; Photorhabdus; immunity; thioester-containing protein.
Figures



Comment in
-
Thioester-containing proteins: At the crossroads of immune effector mechanisms.Virulence. 2017 Nov 17;8(8):1468-1470. doi: 10.1080/21505594.2017.1355662. Epub 2017 Aug 8. Virulence. 2017. PMID: 28704162 Free PMC article. No abstract available.
References
-
- Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol 2007; 25:697-743; PMID:17201680; https://doi.org/ 10.1146/annurev.immunol.25.022106.14161510.1146/annurev.immunol.25.022106.141615 - DOI - PubMed
-
- Pal S, Wu LP. Pattern recognition receptors in the fly: lessons we can learn from the Drosophila melanogaster immune system. Fly (Austin) 2009; 3:121-9; PMID:19440043; https://doi.org/ 10.4161/fly.882710.4161/fly.8827 - DOI - PubMed
-
- Buresova V, Hajdusek O, Franta Z, Sojka D, Kopacek P. IrAM-An alpha2-macroglobulin from the hard tick Ixodes ricinus: characterization and function in phagocytosis of a potential pathogen Chryseobacterium indologenes. Dev Comp Immunol 2009; 33:489-98; PMID:18948134; https://doi.org/ 10.1016/j.dci.2008.09.01110.1016/j.dci.2008.09.011 - DOI - PubMed
-
- Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 2004; 116:661-70; PMID:15006349; https://doi.org/ 10.1016/S0092-8674(04)00173-410.1016/S0092-8674(04)00173-4 - DOI - PubMed
-
- Xiao X, Liu Y, Zhang X, Wang J, Li Z, Pang X, Wang P, Cheng G. Complement-related proteins control the flavivirus infection of Aedes aegypti by inducing antimicrobial peptides. PLoS Pathog 2014; 10:e1004027; PMID:24722701; https://doi.org/ 10.1371/journal.ppat.100402710.1371/journal.ppat.1004027 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
