Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation
- PMID: 28498833
- PMCID: PMC5428915
- DOI: 10.1371/journal.pone.0176280
Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation
Abstract
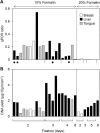
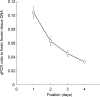
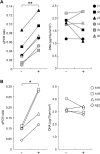
Formalin-fixed, paraffin-embedded (FFPE) tissues used for pathological diagnosis are valuable for studying cancer genomics. In particular, laser-capture microdissection of target cells determined by histopathology combined with FFPE tissue section immunohistochemistry (IHC) enables precise analysis by next-generation sequencing (NGS) of the genetic events occurring in cancer. The result is a new strategy for a pathological tool for cancer diagnosis: 'microgenomics'. To more conveniently and precisely perform microgenomics, we revealed by systematic analysis the following three details regarding FFPE DNA compared with paired frozen tissue DNA. 1) The best quality of FFPE DNA is obtained by tissue fixation with 10% neutral buffered formalin for 1 day and heat treatment of tissue lysates at 95°C for 30 minutes. 2) IHC staining of FFPE tissues decreases the quantity and quality of FFPE DNA to one-fourth, and antigen retrieval (at 120°C for 15 minutes, pH 6.0) is the major reason for this decrease. 3) FFPE DNA prepared as described herein is sufficient for NGS. For non-mutated tissue specimens, no artifactual mutation occurs during FFPE preparation, as shown by precise comparison of NGS of FFPE DNA and paired frozen tissue DNA followed by validation. These results demonstrate that even FFPE tissues used for routine clinical diagnosis can be utilized to obtain reliable NGS data if appropriate conditions of fixation and validation are applied.
Conflict of interest statement
Figures





References
-
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science. 2013; 339(6127): 1546–58. PubMed Central PMCID: PMC3749880. doi: 10.1126/science.1235122 - DOI - PMC - PubMed
-
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013; 153(1): 17–37. doi: 10.1016/j.cell.2013.03.002 - DOI - PubMed
-
- Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015; 61(1): 64–71. doi: 10.1373/clinchem.2014.223040 - DOI - PubMed
-
- Nakayama Y, Yamaguchi H, Einaga N, Esumi M. Pitfalls of DNA Quantification Using DNA-Binding Fluorescent Dyes and Suggested Solutions. PLoS One. 2016; 11(3): e0150528 PubMed Central PMCID: PMC4777359. doi: 10.1371/journal.pone.0150528 - DOI - PMC - PubMed
-
- Wong SQ, Li J, Tan AY, Vedururu R, Pang JM, Do H, et al. Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med Genomics. 2014; 7: 23 PubMed Central PMCID: PMC4032349. doi: 10.1186/1755-8794-7-23 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

