Mycobacterium tuberculosis Whole Genome Sequences From Southern India Suggest Novel Resistance Mechanisms and the Need for Region-Specific Diagnostics
- PMID: 28498943
- PMCID: PMC5434337
- DOI: 10.1093/cid/cix169
Mycobacterium tuberculosis Whole Genome Sequences From Southern India Suggest Novel Resistance Mechanisms and the Need for Region-Specific Diagnostics
Erratum in
-
Erratum.Clin Infect Dis. 2017 Sep 15;65(6):1057. doi: 10.1093/cid/cix522. Clin Infect Dis. 2017. PMID: 28903512 Free PMC article. No abstract available.
-
Erratum.Clin Infect Dis. 2017 Oct 15;65(8):1431-1433. doi: 10.1093/cid/cix563. Clin Infect Dis. 2017. PMID: 29017252 Free PMC article. No abstract available.
-
Correction for 87482 - cix169.Clin Infect Dis. 2024 Dec 17;79(6):1544. doi: 10.1093/cid/ciae505. Clin Infect Dis. 2024. PMID: 39545814 Free PMC article. No abstract available.
Abstract
Background.: India is home to 25% of all tuberculosis cases and the second highest number of multidrug resistant cases worldwide. However, little is known about the genetic diversity and resistance determinants of Indian Mycobacterium tuberculosis, particularly for the primary lineages found in India, lineages 1 and 3.
Methods.: We whole genome sequenced 223 randomly selected M. tuberculosis strains from 196 patients within the Tiruvallur and Madurai districts of Tamil Nadu in Southern India. Using comparative genomics, we examined genetic diversity, transmission patterns, and evolution of resistance.
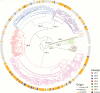
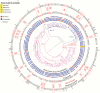
Results.: Genomic analyses revealed (11) prevalence of strains from lineages 1 and 3, (11) recent transmission of strains among patients from the same treatment centers, (11) emergence of drug resistance within patients over time, (11) resistance gained in an order typical of strains from different lineages and geographies, (11) underperformance of known resistance-conferring mutations to explain phenotypic resistance in Indian strains relative to studies focused on other geographies, and (11) the possibility that resistance arose through mutations not previously implicated in resistance, or through infections with multiple strains that confound genotype-based prediction of resistance.
Conclusions.: In addition to substantially expanding the genomic perspectives of lineages 1 and 3, sequencing and analysis of M. tuberculosis whole genomes from Southern India highlight challenges of infection control and rapid diagnosis of resistant tuberculosis using current technologies. Further studies are needed to fully explore the complement of diversity and resistance determinants within endemic M. tuberculosis populations.
Keywords: CAS lineage; EAI lineage; India; Indo-Oceanic lineage; drug resistance..
© The Author 2017. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures
Comment in
-
Reply to Lee and Howden.Clin Infect Dis. 2018 Jan 6;66(1):160-161. doi: 10.1093/cid/cix751. Clin Infect Dis. 2018. PMID: 29040415 Free PMC article. No abstract available.
-
The Critical Importance of Sampling Fraction to Inferences of Mycobacterium tuberculosis Transmission.Clin Infect Dis. 2018 Jan 6;66(1):159-160. doi: 10.1093/cid/cix750. Clin Infect Dis. 2018. PMID: 29040477 Free PMC article. No abstract available.
-
Limited Evidence for the Need for Region-Specific, Genotypic Drug-Susceptibility Assays for Mycobacterium tuberculosis.Clin Infect Dis. 2018 Apr 17;66(9):1481-1482. doi: 10.1093/cid/cix1042. Clin Infect Dis. 2018. PMID: 29182771 No abstract available.
References
-
- WHO. Global Tuberculosis Report 2016. World Health Organization; 2016.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

