High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system
- PMID: 28499063
- PMCID: PMC5785356
- DOI: 10.1111/pbi.12755
High efficient multisites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system
Abstract
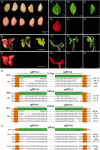
Gossypium hirsutum is an allotetraploid with a complex genome. Most genes have multiple copies that belong to At and Dt subgenomes. Sequence similarity is also very high between gene homologues. To efficiently achieve site/gene-specific mutation is quite needed. Due to its high efficiency and robustness, the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 system has exerted broad site-specific genome editing from prokaryotes to eukaryotes. In this study, we utilized a CRISPR/Cas9 system to generate two sgRNAs in a single vector to conduct multiple sites genome editing in allotetraploid cotton. An exogenously transformed gene Discosoma red fluorescent protein2(DsRed2) and an endogenous gene GhCLA1 were chosen as targets. The DsRed2-edited plants in T0 generation reverted its traits to wild type, with vanished red fluorescence the whole plants. Besides, the mutated phenotype and genotype were inherited to their T1 progenies. For the endogenous gene GhCLA1, 75% of regenerated plants exhibited albino phenotype with obvious nucleotides and DNA fragments deletion. The efficiency of gene editing at each target site is 66.7-100%. The mutation genotype was checked for both genes with Sanger sequencing. Barcode-based high-throughput sequencing, which could be highly efficient for genotyping to a population of mutants, was conducted in GhCLA1-edited T0 plants and it matched well with Sanger sequencing results. No off-target editing was detected at the potential off-target sites. These results prove that the CRISPR/Cas9 system is highly efficient and reliable for allotetraploid cotton genome editing.
Keywords: CRISPR/Cas9; allotetraploid; cotton; genome editing; high-throughput sequencing.
© 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures





References
-
- Bhaya, D. , Davison, M. and Barrangou, R. (2011) CRISPR‐Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

