Impaired Mitochondrial Transcription Termination Disrupts the Stromal Redox Poise in Chlamydomonas
- PMID: 28500267
- PMCID: PMC5490881
- DOI: 10.1104/pp.16.00946
Impaired Mitochondrial Transcription Termination Disrupts the Stromal Redox Poise in Chlamydomonas
Abstract
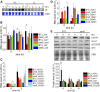
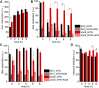
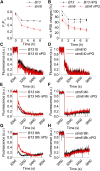
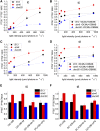
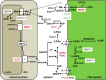
In photosynthetic eukaryotes, the metabolite exchange between chloroplast and mitochondria ensures efficient photosynthesis under saturating light conditions. The Chlamydomonas reinhardtii mutant stm6 is devoid of the mitochondrial transcription termination factor MOC1 and aberrantly expresses the mitochondrial genome, resulting in enhanced photosynthetic hydrogen production and diminished light tolerance. We analyzed the modulation of mitochondrial and chlororespiration during the acclimation of stm6 and the MOC1-complemented strain to excess light. Although light stress stimulated mitochondrial respiration via the energy-conserving cytochrome c pathway in both strains, the mutant was unable to fine-tune the expression and activity of oxidative phosphorylation complex I in excess light, which was accompanied by an increased mitochondrial respiration via the alternative oxidase pathway. Furthermore, stm6 failed to fully activate chlororespiration and cyclic electron flow due to a more oxidized state of the chloroplast stroma, which is caused by an increased mitochondrial electron sink capacity. Increased susceptibility to photoinhibition of PSII in stm6 demonstrates that the MOC1-dependent modulation of mitochondrial respiration helps control the stromal redox poise as a crucial part of high-light acclimation in C. reinhardtii.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Alric J, Lavergne J, Rappaport F (2010) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii (I) aerobic conditions. Biochim Biophys Acta 1797: 44–51 - PubMed
-
- Atteia A, Adrait A, Brugière S, Tardif M, van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W, et al. (2009) A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol Biol Evol 26: 1533–1548 - PubMed
-
- Bailleul B, Berne N, Murik O, Petroutsos D, Prihoda J, Tanaka A, Villanova V, Bligny R, Flori S, Falconet D, et al. (2015) Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369 - PubMed
-
- Bailleul B, Cardol P, Breyton C, Finazzi G (2010) Electrochromism: a useful probe to study algal photosynthesis. Photosynth Res 106: 179–189 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

