A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis
- PMID: 28500268
- PMCID: PMC5490923
- DOI: 10.1104/pp.17.00542
A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis
Abstract
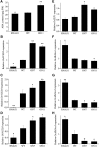
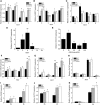
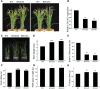
It is well known that abscisic acid (ABA)-induced leaf senescence and premature leaf senescence negatively affect the yield of rice (Oryza sativa). However, the molecular mechanism underlying this relationship, especially the upstream transcriptional network that modulates ABA level during leaf senescence, remains largely unknown. Here, we demonstrate a rice NAC transcription factor, OsNAC2, that participates in ABA-induced leaf senescence. Overexpression of OsNAC2 dramatically accelerated leaf senescence, whereas its knockdown lines showed a delay in leaf senescence. Chromatin immunoprecipitation-quantitative PCR, dual-luciferase, and yeast one-hybrid assays demonstrated that OsNAC2 directly activates expression of chlorophyll degradation genes, OsSGR and OsNYC3 Moreover, ectopic expression of OsNAC2 leads to an increase in ABA levels via directly up-regulating expression of ABA biosynthetic genes (OsNCED3 and OsZEP1) as well as down-regulating the ABA catabolic gene (OsABA8ox1). Interestingly, OsNAC2 is upregulated by a lower level of ABA but downregulated by a higher level of ABA, indicating a feedback repression of OsNAC2 by ABA. Additionally, reduced OsNAC2 expression leads to about 10% increase in the grain yield of RNAi lines. The novel ABA-NAC-SAGs regulatory module might provide a new insight into the molecular action of ABA to enhance leaf senescence and elucidates the transcriptional network of ABA production during leaf senescence in rice.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




Similar articles
-
OsWRKY5 Promotes Rice Leaf Senescence via Senescence-Associated NAC and Abscisic Acid Biosynthesis Pathway.Int J Mol Sci. 2019 Sep 9;20(18):4437. doi: 10.3390/ijms20184437. Int J Mol Sci. 2019. PMID: 31505875 Free PMC article.
-
OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice.Proc Natl Acad Sci U S A. 2014 Jul 8;111(27):10013-8. doi: 10.1073/pnas.1321568111. Epub 2014 Jun 20. Proc Natl Acad Sci U S A. 2014. PMID: 24951508 Free PMC article.
-
Multilayered Regulation of Membrane-Bound ONAC054 Is Essential for Abscisic Acid-Induced Leaf Senescence in Rice.Plant Cell. 2020 Mar;32(3):630-649. doi: 10.1105/tpc.19.00569. Epub 2020 Jan 6. Plant Cell. 2020. PMID: 31911455 Free PMC article.
-
Current Understanding of Leaf Senescence in Rice.Int J Mol Sci. 2021 Apr 26;22(9):4515. doi: 10.3390/ijms22094515. Int J Mol Sci. 2021. PMID: 33925978 Free PMC article. Review.
-
Abiotic Stresses Intervene with ABA Signaling to Induce Destructive Metabolic Pathways Leading to Death: Premature Leaf Senescence in Plants.Int J Mol Sci. 2019 Jan 10;20(2):256. doi: 10.3390/ijms20020256. Int J Mol Sci. 2019. PMID: 30634648 Free PMC article. Review.
Cited by
-
YELLOW LEAF AND DWARF 7, Encoding a Novel Ankyrin Domain-Containing Protein, Affects Chloroplast Development in Rice.Genes (Basel). 2024 Sep 27;15(10):1267. doi: 10.3390/genes15101267. Genes (Basel). 2024. PMID: 39457391 Free PMC article.
-
Mutation of Leaf Senescence 1 Encoding a C2H2 Zinc Finger Protein Induces ROS Accumulation and Accelerates Leaf Senescence in Rice.Int J Mol Sci. 2022 Nov 21;23(22):14464. doi: 10.3390/ijms232214464. Int J Mol Sci. 2022. PMID: 36430940 Free PMC article.
-
The auxin response factor TaARF15-A1 negatively regulates senescence in common wheat (Triticum aestivum L.).Plant Physiol. 2023 Feb 12;191(2):1254-1271. doi: 10.1093/plphys/kiac497. Plant Physiol. 2023. PMID: 36282536 Free PMC article.
-
Integrated transcriptomics and miRNAomics provide insights into the complex multi-tiered regulatory networks associated with coleoptile senescence in rice.Front Plant Sci. 2022 Oct 12;13:985402. doi: 10.3389/fpls.2022.985402. eCollection 2022. Front Plant Sci. 2022. PMID: 36311124 Free PMC article.
-
Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways.Rice (N Y). 2019 Oct 21;12(1):76. doi: 10.1186/s12284-019-0334-6. Rice (N Y). 2019. PMID: 31637532 Free PMC article.
References
-
- Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62: 250–264 - PubMed
-
- Becker W, Apel K (1993) Differences in gene expression between natural and artificially induced leaf senescence. Planta 189: 74–79
-
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18: 756–767 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

