Network and role analysis of autophagy in Phytophthora sojae
- PMID: 28500315
- PMCID: PMC5431975
- DOI: 10.1038/s41598-017-01988-7
Network and role analysis of autophagy in Phytophthora sojae
Abstract
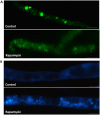
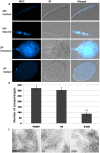
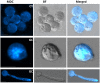
Autophagy is an evolutionarily conserved mechanism in eukaryotes with roles in development and the virulence of plant fungal pathogens. However, few reports on autophagy in oomycete species have been published. Here, we identified 26 autophagy-related genes (ATGs) belonging to 20 different groups in Phytophthora sojae using a genome-wide survey, and core ATGs in oomycetes were used to construct a preliminary autophagy pathway model. Expression profile analysis revealed that these ATGs are broadly expressed and that the majority of them significantly increase during infection stages, suggesting a central role for autophagy in virulence. Autophagy in P. sojae was detected using a GFP-PsAtg8 fusion protein and the fluorescent dye MDC during rapamycin and starvation treatment. In addition, autophagy was significantly induced during sporangium formation and cyst germination. Silencing PsAtg6a in P. sojae significantly reduced sporulation and pathogenicity. Furthermore, a PsAtg6a-silenced strain showed haustorial formation defects. These results suggested that autophagy might play essential roles in both the development and infection mechanism of P. sojae.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Phylogenetic and transcriptional analysis of an expanded bZIP transcription factor family in Phytophthora sojae.BMC Genomics. 2013 Nov 28;14(1):839. doi: 10.1186/1471-2164-14-839. BMC Genomics. 2013. PMID: 24286285 Free PMC article.
-
Phytophthora sojae TatD nuclease positively regulates sporulation and negatively regulates pathogenesis.Mol Plant Microbe Interact. 2014 Oct;27(10):1070-80. doi: 10.1094/MPMI-05-14-0153-R. Mol Plant Microbe Interact. 2014. PMID: 24940989
-
The Pectin Methylesterase Gene Complement of Phytophthora sojae: Structural and Functional Analyses, and the Evolutionary Relationships with Its Oomycete Homologs.PLoS One. 2015 Nov 6;10(11):e0142096. doi: 10.1371/journal.pone.0142096. eCollection 2015. PLoS One. 2015. PMID: 26544849 Free PMC article.
-
Phytophthora genomics: the plant destroyers' genome decoded.Mol Plant Microbe Interact. 2006 Dec;19(12):1295-301. doi: 10.1094/MPMI-19-1295. Mol Plant Microbe Interact. 2006. PMID: 17153913 Review.
-
Phytopathogenic oomycetes: a review focusing on Phytophthora cinnamomi and biotechnological approaches.Mol Biol Rep. 2020 Nov;47(11):9179-9188. doi: 10.1007/s11033-020-05911-8. Epub 2020 Oct 17. Mol Biol Rep. 2020. PMID: 33068230 Review.
Cited by
-
TOR Inhibitors Synergistically Suppress the Growth and Development of Phytophthora infestans, a Highly Destructive Pathogenic Oomycete.Front Microbiol. 2021 Apr 16;12:596874. doi: 10.3389/fmicb.2021.596874. eCollection 2021. Front Microbiol. 2021. PMID: 33935983 Free PMC article.
-
Evolutionary conservation and metabolic significance of autophagy in algae.Philos Trans R Soc Lond B Biol Sci. 2024 Nov 18;379(1914):20230368. doi: 10.1098/rstb.2023.0368. Epub 2024 Sep 30. Philos Trans R Soc Lond B Biol Sci. 2024. PMID: 39343016 Free PMC article. Review.
-
Deciphering the Biomolecules from Bacillus atrophaeus NMB01 Untangles the Anti-Oomycetes Action of Trioxsalen and Corynan-17-ol, Against Phytophthora infestans Inciting Late Blight of Potato.Indian J Microbiol. 2022 Dec;62(4):641-650. doi: 10.1007/s12088-022-01044-7. Epub 2022 Nov 2. Indian J Microbiol. 2022. PMID: 36458213 Free PMC article.
-
Current opinions on autophagy in pathogenicity of fungi.Virulence. 2019 Dec;10(1):481-489. doi: 10.1080/21505594.2018.1551011. Epub 2018 Dec 3. Virulence. 2019. PMID: 30475080 Free PMC article. Review.
-
Autophagy-Related Gene PlATG6a Is Involved in Mycelial Growth, Asexual Reproduction and Tolerance to Salt and Oxidative Stresses in Peronophythora litchii.Int J Mol Sci. 2022 Feb 6;23(3):1839. doi: 10.3390/ijms23031839. Int J Mol Sci. 2022. PMID: 35163762 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

