Novel Algorithms for Improved Sensitivity in Non-Invasive Prenatal Testing
- PMID: 28500333
- PMCID: PMC5431782
- DOI: 10.1038/s41598-017-02031-5
Novel Algorithms for Improved Sensitivity in Non-Invasive Prenatal Testing
Abstract
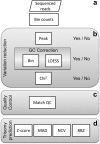
Non-invasive prenatal testing (NIPT) of cell-free DNA in maternal plasma, which is a mixture of maternal DNA and a low percentage of fetal DNA, can detect fetal aneuploidies using massively parallel sequencing. Because of the low percentage of fetal DNA, methods with high sensitivity and precision are required. However, sequencing variation lowers sensitivity and hampers detection of trisomy samples. Therefore, we have developed three algorithms to improve sensitivity and specificity: the chi-squared-based variation reduction (χ2VR), the regression-based Z-score (RBZ) and the Match QC score. The χ2VR reduces variability in sequence read counts per chromosome between samples, the RBZ allows for more precise trisomy prediction, and the Match QC score shows if the control group used is representative for a specific sample. We compared the performance of χ2VR to that of existing variation reduction algorithms (peak and GC correction) and that of RBZ to trisomy prediction algorithms (standard Z-score, normalized chromosome value and median-absolute-deviation-based Z-score). χ2VR and the RBZ both reduce variability more than existing methods, and thereby increase the sensitivity of the NIPT analysis. We found the optimal combination of algorithms was to use both GC correction and χ2VR for pre-processing and to use RBZ as the trisomy prediction method.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

