Identification and characterization of a grain micronutrient-related OsFRO2 rice gene ortholog from micronutrient-rich little millet (Panicum sumatrense)
- PMID: 28500402
- PMCID: PMC5429308
- DOI: 10.1007/s13205-017-0656-2
Identification and characterization of a grain micronutrient-related OsFRO2 rice gene ortholog from micronutrient-rich little millet (Panicum sumatrense)
Abstract
Minor millets are considered as nutrient-rich cereals having significant effect in improving human health. In this study, a rice ortholog of Ferric Chelate Reductase (FRO2) gene involved in plant metal uptake has been identified in iron-rich Little millet (LM) using PCR and next generation sequencing-based strategy. FRO2 gene-specific primers designed from rice genome amplified 2.7 Kb fragment in LM genotype RLM-37. Computational genomics analyses of the sequenced amplicon showed high level sequence similarity with rice OsFRO2 gene. The predicted gene structure showed the presence of 6 exons and 5 introns and its protein sequence was found to contain ferric reductase and NOX_Duox_Like_FAD_NADP domains. Further, 3D structure analysis of FCR-LM model protein (494 amino acids) shows that it has 18 helices, 10 beta sheets, 10 strands, 41 beta turn and 5 gamma turn with slight deviation from the FCR-Os structure. Besides, the structures of FCR-LM and FCR-Os were modelled followed by molecular dynamics simulations. The overall study revealed both sequence and structural similarity between the identified gene and OsFRO2. Thus, a putative ferric chelate reductase gene has been identified in LM paving the way for using this approach for identification of orthologs of other metal genes from millets. This also facilitates mining of effective alleles of known genes for improvement of staple crops like rice.
Keywords: Ferric Chelate Reductase; Little millet; MD simulation; Metal homeostasis; Sequencing.
Conflict of interest statement
The authors do not have any conflict of interest.
Figures
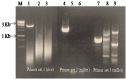


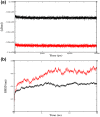


References
-
- Amadou I, Gounga ME, Le GW. Millets: nutritional composition, some health benefits and processing—A review. Emir J Food Agric. 2013;25:501–508. doi: 10.9755/ejfa.v25i7.12045. - DOI
-
- Berendsen HJC, Postma JPM, Van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. In: Pullman B, editor. Intermolecular forces. Dordrecht: Reidel; 1981. pp. 331–342.
-
- Berendsen HJC, Postma JPM, Van Gunsteren WF, Dinola A, Haak JR. Molecular Dynamic with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. doi: 10.1063/1.448118. - DOI
-
- Berk H, Carsten K, David VS, Erik L. GROMACS 4: algorithms for highly efficient, load balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):335–447. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

