Stability of Proteins in Dried Blood Spot Biobanks
- PMID: 28501802
- PMCID: PMC5500761
- DOI: 10.1074/mcp.RA117.000015
Stability of Proteins in Dried Blood Spot Biobanks
Abstract
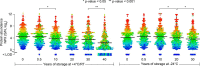
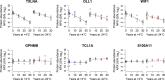
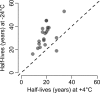
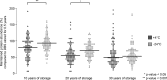
An important motivation for the construction of biobanks is to discover biomarkers that identify diseases at early, potentially curable stages. This will require biobanks from large numbers of individuals, preferably sampled repeatedly, where the samples are collected and stored under conditions that preserve potential biomarkers. Dried blood samples are attractive for biobanking because of the ease and low cost of collection and storage. Here we have investigated their suitability for protein measurements. Ninety-two proteins with relevance for oncology were analyzed using multiplex proximity extension assays (PEA) in dried blood spots collected on paper and stored for up to 30 years at either +4 °C or -24 °C.Our main findings were that (1) the act of drying only slightly influenced detection of blood proteins (average correlation of 0.970), and in a reproducible manner (correlation of 0.999), (2) detection of some proteins was not significantly affected by storage over the full range of three decades (34 and 76% of the analyzed proteins at +4 °C and -24 °C, respectively), whereas levels of others decreased slowly during storage with half-lives in the range of 10 to 50 years, and (3) detectability of proteins was less affected in dried samples stored at -24 °C compared with at +4 °C, as the median protein abundance had decreased to 80 and 93% of starting levels after 10 years of storage at +4 °C or -24 °C, respectively. The results of our study are encouraging as they suggest an inexpensive means to collect large numbers of blood samples, even by the donors themselves, and to transport, and store biobanked samples as spots of whole blood dried on paper. Combined with emerging means to measure hundreds or thousands of protein, such biobanks could prove of great medical value by greatly enhancing discovery as well as routine analysis of blood biomarkers.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
Competing interests: U.L. is a founder of and holds stock in Olink Proteomics commercializing the PEA panels used herein
Figures

References
-
- Ignjatovic V., Pitt J., Monagle P., and Craig J. M. (2014) The utility of dried blood spots for proteomic studies: looking forward to looking back. Proteomics Clin. Appl. 8, 896–900 - PubMed
-
- McDade T. W., Williams S., and Snodgrass J. J. (2007) What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography 44, 899–925 - PubMed
-
- Razavi M., Anderson N. L., Yip R., Pope M. E., and Pearson T. W. (2016) Multiplexed longitudinal measurement of protein biomarkers in DBS using an automated SISCAPA workflow. Bioanalysis 8, 1597–1609 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

