Gene co-expression networks identify Trem2 and Tyrobp as major hubs in human APOE expressing mice following traumatic brain injury
- PMID: 28502803
- PMCID: PMC5530876
- DOI: 10.1016/j.nbd.2017.05.006
Gene co-expression networks identify Trem2 and Tyrobp as major hubs in human APOE expressing mice following traumatic brain injury
Abstract
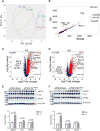
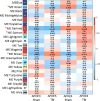
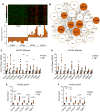
Traumatic brain injury (TBI) is strongly linked to an increased risk of developing dementia, including chronic traumatic encephalopathy and possibly Alzheimer's disease (AD). APOEε4 allele of human Apolipoprotein E (APOE) gene is the major genetic risk factor for late onset AD and has been associated with chronic traumatic encephalopathy and unfavorable outcome following TBI. To determine if there is an APOE isoform-specific response to TBI we performed controlled cortical impact on 3-month-old mice expressing human APOE3 or APOE4 isoforms. Following injury, we used several behavior paradigms to test for anxiety and learning and found that APOE3 and APOE4 targeted replacement mice demonstrate cognitive impairments following moderate TBI. Transcriptional profiling 14days following injury revealed a significant effect of TBI, which was similar in both genotypes. Significantly upregulated by injury in both genotypes were mRNA expression and protein level of ABCA1 transporter and APOJ, but not APOE. To identify gene-networks correlated to injury and APOE isoform, we performed Weighted Gene Co-expression Network Analysis. We determined that the network mostly correlated to TBI in animals expressing both isoforms is immune response with major hub genes including Trem2, Tyrobp, Clec7a and Cd68. We also found a significant increase of TREM2, IBA-1 and GFAP protein levels in the brains of injured mice. We identified a network representing myelination that correlated significantly with APOE isoform in both injury groups. This network was significantly enriched in oligodendrocyte signature genes, such as Mbp and Plp1. Our results demonstrate unique and distinct gene networks at this acute time point for injury and APOE isoform, as well as a network driven by APOE isoform across TBI groups.
Keywords: Apolipoprotein E; Fyn; Immune response; Innate immune response; Myelination; Traumatic brain injury; Trem2; Tyrobp.
Copyright © 2017 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest: The authors declare that they have no conflict of interest.
Figures





References
-
- Abraham CR. Reactive astrocytes and alpha1-antichymotrypsin in Alzheimer’s disease. Neurobiol Aging. 2001;22:931–936. - PubMed
-
- Acosta SA, Tajiri N, Shinozuka K, Ishikawa H, Grimmig B, Diamond DM, Diamond D, Sanberg PR, Bickford PC, Kaneko Y, Borlongan CV. Long-term upregulation of inflammation and suppression of cell proliferation in the brain of adult rats exposed to traumatic brain injury using the controlled cortical impact model. PloS one. 2013;8:e53376–e53376. - PMC - PubMed
-
- Alderson NL, Maldonado EN, Kern MJ, Bhat NR, Hama H. FA2H-dependent fatty acid 2-hydroxylation in postnatal mouse brain. J Lipid Res. 2006;47:2772–2780. - PubMed
-
- Alexander S, Kerr ME, Kim Y, Kamboh MI. Apolipoprotein E4 allele presence and functional outcome after severe traumatic brain injury. Journal of Neurotrauma. 2007;24:790–797. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

