Drosophila melanogaster as a High-Throughput Model for Host-Microbiota Interactions
- PMID: 28503170
- PMCID: PMC5408076
- DOI: 10.3389/fmicb.2017.00751
Drosophila melanogaster as a High-Throughput Model for Host-Microbiota Interactions
Abstract
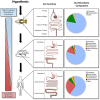
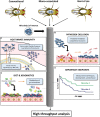
Microbiota research often assumes that differences in abundance and identity of microorganisms have unique influences on host physiology. To test this concept mechanistically, germ-free mice are colonized with microbial communities to assess causation. Due to the cost, infrastructure challenges, and time-consuming nature of germ-free mouse models, an alternative approach is needed to investigate host-microbial interactions. Drosophila melanogaster (fruit flies) can be used as a high throughput in vivo screening model of host-microbiome interactions as they are affordable, convenient, and replicable. D. melanogaster were essential in discovering components of the innate immune response to pathogens. However, axenic D. melanogaster can easily be generated for microbiome studies without the need for ethical considerations. The simplified microbiota structure enables researchers to evaluate permutations of how each microbial species within the microbiota contribute to host phenotypes of interest. This enables the possibility of thorough strain-level analysis of host and microbial properties relevant to physiological outcomes. Moreover, a wide range of mutant D. melanogaster strains can be affordably obtained from public stock centers. Given this, D. melanogaster can be used to identify candidate mechanisms of host-microbe symbioses relevant to pathogen exclusion, innate immunity modulation, diet, xenobiotics, and probiotic/prebiotic properties in a high throughput manner. This perspective comments on the most promising areas of microbiota research that could immediately benefit from using the D. melanogaster model.
Keywords: Drosophila melanogaster; animal model; fruit fly; microbiome; microbiota; prebiotic; probiotic; symbiosis.
Figures
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

