HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells
- PMID: 28504668
- PMCID: PMC5548598
- DOI: 10.1038/nbt.3860
HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells
Abstract
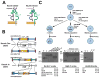
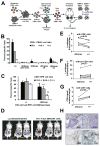
Polymorphisms in the human leukocyte antigen (HLA) class I genes can cause the rejection of pluripotent stem cell (PSC)-derived products in allogeneic recipients. Disruption of the Beta-2 Microglobulin (B2M) gene eliminates surface expression of all class I molecules, but leaves the cells vulnerable to lysis by natural killer (NK) cells. Here we show that this 'missing-self' response can be prevented by forced expression of minimally polymorphic HLA-E molecules. We use adeno-associated virus (AAV)-mediated gene editing to knock in HLA-E genes at the B2M locus in human PSCs in a manner that confers inducible, regulated, surface expression of HLA-E single-chain dimers (fused to B2M) or trimers (fused to B2M and a peptide antigen), without surface expression of HLA-A, B or C. These HLA-engineered PSCs and their differentiated derivatives are not recognized as allogeneic by CD8+ T cells, do not bind anti-HLA antibodies and are resistant to NK-mediated lysis. Our approach provides a potential source of universal donor cells for applications where the differentiated derivatives lack HLA class II expression.
Figures


Comment in
-
Pluripotent stem cells that evade the immune radar.Nat Biotechnol. 2017 Aug 8;35(8):722-723. doi: 10.1038/nbt.3940. Nat Biotechnol. 2017. PMID: 28787417 No abstract available.
References
-
- Taylor CJ, Peacock S, Chaudhry AN, Bradley JA, Bolton EM. Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient hla types. Cell Stem Cell. 2012;11:147–152. - PubMed
-
- Nishikawa S, Goldstein Ra, Nierras CR. The promise of human induced pluripotent stem cells for research and therapy. Nat Rev Mol Cell Biol. 2008;9:725–729. - PubMed
-
- Taylor CJ, et al. Banking on human embryonic stem cells: Estimating the number of donor cell lines needed for HLA matching. Lancet. 2005;366:2019–2025. - PubMed
-
- Braciale TJ. Antigen processing for presentation by MHC class I molecules. Curr Opin Immunol. 1992;4:59–62. - PubMed
-
- Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF. The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin. Tissue Antigens. 1978;11:96–112. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

