Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation
- PMID: 28504689
- PMCID: PMC5523069
- DOI: 10.1038/oncsis.2017.36
Molecular signaling in multiple myeloma: association of RAS/RAF mutations and MEK/ERK pathway activation
Abstract
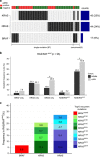
Multiple myeloma (MM) is a plasma cell malignancy that is still considered to be incurable in most cases. A dominant mutation cluster has been identified in RAS/RAF genes, emphasizing the potential significance of RAS/RAF/MEK/ERK signaling as a therapeutic target. As yet, however, the clinical relevance of this finding is unclear as clinical responses to MEK inhibition in RAS-mutant MM have been mixed. We therefore assessed RAS/RAF mutation status and MEK/ERK pathway activation by both targeted sequencing and phospho-ERK immunohistochemistry in 180 tissue biopsies from 103 patients with newly diagnosed MM (NDMM) and 77 patients with relapsed/refractory MM (rrMM). We found a significant enrichment of RAS/BRAF mutations in rrMM compared to NDMM (P=0.011), which was mainly due to an increase of NRAS mutations (P=0.010). As expected, BRAF mutations were significantly associated with activated downstream signaling. However, only KRAS and not NRAS mutations were associated with pathway activation compared to RAS/BRAFwt (P=0.030). More specifically, only KRASG12D and BRAFV600E were consistently associated with ERK activation (P<0.001 and P=0.006, respectively). Taken together, these results suggest the need for a more specific stratification strategy consisting of both confirmation of protein-level pathway activation as well as detailed RAS/RAF mutation status to allow for a more precise and more effective application of targeted therapies, for example, with BRAF/MEK inhibitors in MM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mandal R, Becker S, Strebhardt K. Stamping out RAF and MEK1/2 to inhibit the ERK1/2 pathway: an emerging threat to anticancer therapy. Oncogene 2016; 35: 2547–2561. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

