The mitochondrial respiratory chain is essential for haematopoietic stem cell function
- PMID: 28504706
- PMCID: PMC5474760
- DOI: 10.1038/ncb3529
The mitochondrial respiratory chain is essential for haematopoietic stem cell function
Abstract
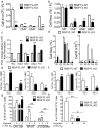
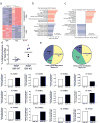
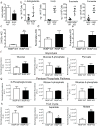
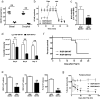
Adult and fetal haematopoietic stem cells (HSCs) display a glycolytic phenotype, which is required for maintenance of stemness; however, whether mitochondrial respiration is required to maintain HSC function is not known. Here we report that loss of the mitochondrial complex III subunit Rieske iron-sulfur protein (RISP) in fetal mouse HSCs allows them to proliferate but impairs their differentiation, resulting in anaemia and prenatal death. RISP-null fetal HSCs displayed impaired respiration resulting in a decreased NAD+/NADH ratio. RISP-null fetal HSCs and progenitors exhibited an increase in both DNA and histone methylation associated with increases in 2-hydroxyglutarate (2HG), a metabolite known to inhibit DNA and histone demethylases. RISP inactivation in adult HSCs also impaired respiration resulting in loss of quiescence concomitant with severe pancytopenia and lethality. Thus, respiration is dispensable for adult or fetal HSC proliferation, but essential for fetal HSC differentiation and maintenance of adult HSC quiescence.
Figures



References
-
- Chandel NS, Jasper H, Ho TT, Passegue E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nature cell biology. 2016;18:823–832. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

