Molecular Characterization and Expression Profiling of Tomato GRF Transcription Factor Family Genes in Response to Abiotic Stresses and Phytohormones
- PMID: 28505092
- PMCID: PMC5454968
- DOI: 10.3390/ijms18051056
Molecular Characterization and Expression Profiling of Tomato GRF Transcription Factor Family Genes in Response to Abiotic Stresses and Phytohormones
Abstract
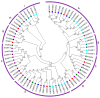
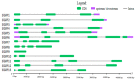
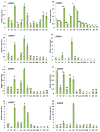
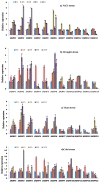
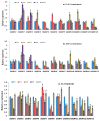
Growth regulating factors (GRFs) are plant-specific transcription factors that are involved in diverse biological and physiological processes, such as growth, development and stress and hormone responses. However, the roles of GRFs in vegetative and reproductive growth, development and stress responses in tomato (Solanum lycopersicum) have not been extensively explored. In this study, we characterized the 13 SlGRF genes. In silico analysis of protein motif organization, intron-exon distribution, and phylogenetic classification confirmed the presence of GRF proteins in tomato. The tissue-specific expression analysis revealed that most of the SlGRF genes were preferentially expressed in young and growing tissues such as flower buds and meristems, suggesting that SlGRFs are important during growth and development of these tissues. Some of the SlGRF genes were preferentially expressed in fruits at distinct developmental stages suggesting their involvement in fruit development and the ripening process. The strong and differential expression of different SlGRFs under NaCl, drought, heat, cold, abscisic acid (ABA), and jasmonic acid (JA) treatment, predict possible functions for these genes in stress responses in addition to their growth regulatory functions. Further, differential expression of SlGRF genes upon gibberellic acid (GA3) treatment indicates their probable function in flower development and stress responses through a gibberellic acid (GA)-mediated pathway. The results of this study provide a basis for further functional analysis and characterization of this important gene family in tomato.
Keywords: GRF gene; Solanum lycopersicum; abiotic stress; fruit development; organ-specific expression; phytohormone treatment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Debernardi J.M., Mecchia M.A., Vercruyssen L., Smaczniak C., Kaufmann K., Inze D., Rodriguez R.E., Palatnik J.F. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 2014;79:413–426. doi: 10.1111/tpj.12567. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

