Structure of Ca2+-binding protein-6 from Entamoeba histolytica and its involvement in trophozoite proliferation regulation
- PMID: 28505197
- PMCID: PMC5444848
- DOI: 10.1371/journal.ppat.1006332
Structure of Ca2+-binding protein-6 from Entamoeba histolytica and its involvement in trophozoite proliferation regulation
Abstract
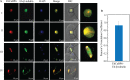
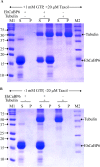
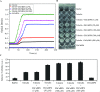
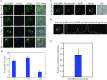
Cell cycle of Entamoeba histolytica, the etiological agent of amoebiasis, follows a novel pathway, which includes nuclear division without the nuclear membrane disassembly. We report a nuclear localized Ca2+-binding protein from E. histolytica (abbreviated hereafter as EhCaBP6), which is associated with microtubules. We determined the 3D solution NMR structure of EhCaBP6, and identified one unusual, one canonical and two non-canonical cryptic EF-hand motifs. The cryptic EF-II and EF-IV pair with the Ca2+-binding EF-I and EF-III, respectively, to form a two-domain structure similar to Calmodulin and Centrin proteins. Downregulation of EhCaBP6 affects cell proliferation by causing delays in transition from G1 to S phase, and inhibition of DNA synthesis and cytokinesis. We also demonstrate that EhCaBP6 modulates microtubule dynamics by increasing the rate of tubulin polymerization. Our results, including structural inferences, suggest that EhCaBP6 is an unusual CaBP involved in regulating cell proliferation in E. histolytica similar to nuclear Calmodulin.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Kahl CR, Means AR (2003) Regulation of cell cycle progression by Calcium/Calmodulin-dependent pathways. Endocr Rev. 24: 719–736. doi: 10.1210/er.2003-0008 - DOI - PubMed
-
- Choi J, Husain M (2006) Calmodulin-mediated cell cycle regulation: new mechanisms for old observation. Cell Cycle. 5: 2183–2186. doi: 10.4161/cc.5.19.3265 - DOI - PubMed
-
- Santella L (1998) The role of calcium in the cell cycle: facts and hypotheses. Biochem Biophys Res Commun. 244: 317–24. doi: 10.1006/bbrc.1998.8086 - DOI - PubMed
-
- Whitaker M, Patel R (1990) Calcium and cell cycle control. Development. 108: 525–542. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

