Phytoplasma-conserved phyllogen proteins induce phyllody across the Plantae by degrading floral MADS domain proteins
- PMID: 28505304
- PMCID: PMC5853863
- DOI: 10.1093/jxb/erx158
Phytoplasma-conserved phyllogen proteins induce phyllody across the Plantae by degrading floral MADS domain proteins
Abstract
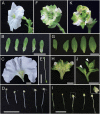
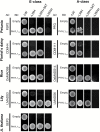
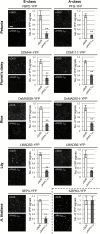
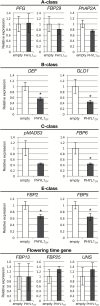
ABCE-class MADS domain transcription factors (MTFs) are key regulators of floral organ development in angiosperms. Aberrant expression of these genes can result in abnormal floral traits such as phyllody. Phyllogen is a virulence factor conserved in phytoplasmas, plant pathogenic bacteria of the class Mollicutes. It triggers phyllody in Arabidopsis thaliana by inducing degradation of A- and E-class MTFs. However, it is still unknown whether phyllogen can induce phyllody in plants other than A. thaliana, although phytoplasma-associated phyllody symptoms are observed in a broad range of angiosperms. In this study, phyllogen was shown to cause phyllody phenotypes in several eudicot species belonging to three different families. Moreover, phyllogen can interact with MTFs of not only angiosperm species including eudicots and monocots but also gymnosperms and a fern, and induce their degradation. These results suggest that phyllogen induces phyllody in angiosperms and inhibits MTF function in diverse plant species.
Keywords: ABCE model; MADS domain transcription factor; floral development; phyllody; phyllogen; phytoplasma.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures

Similar articles
-
Degradation of class E MADS-domain transcription factors in Arabidopsis by a phytoplasmal effector, phyllogen.Plant Signal Behav. 2015;10(8):e1042635. doi: 10.1080/15592324.2015.1042635. Plant Signal Behav. 2015. PMID: 26179462 Free PMC article.
-
Functional variation in phyllogen, a phyllody-inducing phytoplasma effector family, attributable to a single amino acid polymorphism.Mol Plant Pathol. 2020 Oct;21(10):1322-1336. doi: 10.1111/mpp.12981. Epub 2020 Aug 19. Mol Plant Pathol. 2020. PMID: 32813310 Free PMC article.
-
Crystal structure of phyllogen, a phyllody-inducing effector protein of phytoplasma.Biochem Biophys Res Commun. 2019 Jun 11;513(4):952-957. doi: 10.1016/j.bbrc.2019.04.060. Epub 2019 Apr 19. Biochem Biophys Res Commun. 2019. PMID: 31010685
-
Did Convergent Protein Evolution Enable Phytoplasmas to Generate 'Zombie Plants'?Trends Plant Sci. 2015 Dec;20(12):798-806. doi: 10.1016/j.tplants.2015.08.004. Epub 2015 Oct 10. Trends Plant Sci. 2015. PMID: 26463218 Review.
-
MADS-box genes and floral development: the dark side.J Exp Bot. 2012 Sep;63(15):5397-404. doi: 10.1093/jxb/ers233. Epub 2012 Aug 21. J Exp Bot. 2012. PMID: 22915743 Review.
Cited by
-
Plant Hormones in Phytoplasma Infected Plants.Front Plant Sci. 2019 Apr 17;10:477. doi: 10.3389/fpls.2019.00477. eCollection 2019. Front Plant Sci. 2019. PMID: 31057582 Free PMC article. Review.
-
A phytoplasma effector acts as a ubiquitin-like mediator between floral MADS-box proteins and proteasome shuttle proteins.Plant Cell. 2022 Apr 26;34(5):1709-1723. doi: 10.1093/plcell/koac062. Plant Cell. 2022. PMID: 35234248 Free PMC article.
-
Parasitic modulation of host development by ubiquitin-independent protein degradation.Cell. 2021 Sep 30;184(20):5201-5214.e12. doi: 10.1016/j.cell.2021.08.029. Epub 2021 Sep 17. Cell. 2021. PMID: 34536345 Free PMC article.
-
Phytoplasma SAP11 effector destabilization of TCP transcription factors differentially impact development and defence of Arabidopsis versus maize.PLoS Pathog. 2019 Sep 26;15(9):e1008035. doi: 10.1371/journal.ppat.1008035. eCollection 2019 Sep. PLoS Pathog. 2019. PMID: 31557268 Free PMC article.
-
Random mutagenesis-based screening of the interface of phyllogen, a bacterial phyllody-inducing effector, for interaction with plant MADS-box proteins.Front Plant Sci. 2023 Mar 28;14:1058059. doi: 10.3389/fpls.2023.1058059. eCollection 2023. Front Plant Sci. 2023. PMID: 37056494 Free PMC article.
References
-
- Adam H, Jouannic S, Orieux Y, Morcillo F, Richaud F, Duval Y, Tregear JW. 2007. Functional characterization of MADS box genes involved in the determination of oil palm flower structure. Journal of Experimental Botany 58, 1245–1259. - PubMed
-
- Aida R, Komano M, Saito M, Nakase K, Murai K. 2008. Chrysanthemum flower shape modification by suppression of chrysanthemum-AGAMOUS gene. Plant Biotechnology 25, 55–59.
-
- Akhtar KP, Sarwar G, Dickinson M, Ahmad M, Haq MA, Hameed S, Iqbal MJ. 2009. Sesame phyllody disease: its symptomatology, etiology, and transmission in Pakistan. Turkish Journal of Agriculture and Forestry 33, 477–486.
-
- Angenent GC, Franken J, Busscher M, Weiss D, Tunen AJ. 1994. Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. The Plant Journal 5, 33–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

