Rapid susceptibility profiling of carbapenem-resistant Klebsiella pneumoniae
- PMID: 28507322
- PMCID: PMC5432529
- DOI: 10.1038/s41598-017-02009-3
Rapid susceptibility profiling of carbapenem-resistant Klebsiella pneumoniae
Erratum in
-
Author Correction: Rapid susceptibility profiling of carbapenem-resistant Klebsiella pneumoniae.Sci Rep. 2018 Apr 24;8(1):6697. doi: 10.1038/s41598-018-25216-y. Sci Rep. 2018. PMID: 29686361 Free PMC article.
Abstract
The expanding global distribution of multi-resistant Klebsiella pneumoniae demands faster antimicrobial susceptibility testing (AST) to guide antibiotic treatment. Current ASTs rely on time-consuming differentiation of resistance and susceptibility after initial isolation of bacteria from a clinical specimen. Here we describe a flow cytometry workflow to determine carbapenem susceptibility from bacterial cell characteristics in an international K. pneumoniae isolate collection (n = 48), with a range of carbapenemases. Our flow cytometry-assisted susceptibility test (FAST) method combines rapid qualitative susceptible/non-susceptible classification and quantitative MIC measurement in a single process completed shortly after receipt of a primary isolate (54 and 158 minutes respectively). The qualitative FAST results and FAST-derived MIC (MICFAST) correspond closely with broth microdilution MIC (MICBMD, Matthew's correlation coefficient 0.887), align with the international AST standard (ISO 200776-1; 2006) and could be used for rapid determination of antimicrobial susceptibility in a wider range of Gram negative and Gram positive bacteria.
Conflict of interest statement
The corresponding author’s laboratory is funded by a Gates Foundation Grand Challenges award OP1150984 and a Research Translation Project grant from the Health Department of WA. Equipment has been purchased by the Health Department and a matching donation from Rotary Club of Applecross and Lab Without Walls, as stated in the manuscript.
Figures

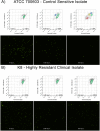

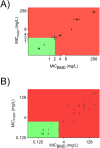


References
-
- The Review on Antimicrobial Resistance. Tackling drug-resistant infections globally: Finalreport and recommendations (ed. O’Neill, J.) 1–72 (HM Government, 2016).
-
- World Health Organization. Antimicrobial resistance. Factsheet no. 194. World Health Organisation Media Centre. http://www.who.int/mediacentre/factsheets/fs194/en/HO AMR challenge (2014).
-
- World Health Organization. WHO’s first global report on antibiotic resistance reveals serious, worldwide threat to public. World Health Organisation Media Centre health, http://www.who.int/mediacentre/news/releases/2014/amr-report/en/ (2014).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

