Distribution of ELOVL4 in the Developing and Adult Mouse Brain
- PMID: 28507511
- PMCID: PMC5410580
- DOI: 10.3389/fnana.2017.00038
Distribution of ELOVL4 in the Developing and Adult Mouse Brain
Abstract
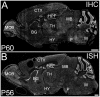
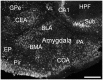
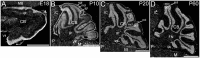
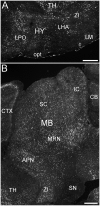
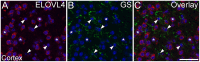
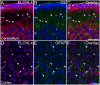
ELOngation of Very Long chain fatty acids (ELOVL)-4 is essential for the synthesis of very long chain-fatty acids (fatty acids with chain lengths ≥ 28 carbons). The functions of ELOVL4 and its very long-chain fatty acid products are poorly understood at present. However, mutations in ELOVL4 cause neurodevelopmental or neurodegenerative diseases that vary according to the mutation and inheritance pattern. Heterozygous inheritance of different ELOVL4 mutations causes Stargardt-like Macular Dystrophy or Spinocerebellar Ataxia type 34. Homozygous inheritance of ELOVL4 mutations causes more severe disease characterized by seizures, intellectual disability, ichthyosis, and premature death. To better understand ELOVL4 and very long chain fatty acid function in the brain, we examined ELOVL4 expression in the mouse brain between embryonic day 18 and postnatal day 60 by immunolabeling using ELOVL4 and other marker antibodies. ELOVL4 was widely expressed in a region- and cell type-specific manner, and was restricted to cell bodies, consistent with its known localization to endoplasmic reticulum. ELOVL4 labeling was most prominent in gray matter, although labeling also was present in some cells located in white matter. ELOVL4 was widely expressed in the developing brain by embryonic day 18 and was especially pronounced in regions underlying the lateral ventricles and other neurogenic regions. The basal ganglia in particular showed intense ELOVL4 labeling at this stage. In the postnatal brain, cerebral cortex, hippocampus, cerebellum, thalamus, hypothalamus, midbrain, pons, and medulla all showed prominent ELOVL4 labeling, although ELOVL4 distribution was not uniform across all cells or subnuclei within these regions. In contrast, the basal ganglia showed little ELOVL4 labeling in the postnatal brain. Double labeling studies showed that ELOVL4 was primarily expressed by neurons, although presumptive oligodendrocytes located in white matter tracts also showed labeling. Little or no ELOVL4 labeling was present in astrocytes or radial glial cells. These findings suggest that ELOVL4 and its very long chain fatty acid products are important in many parts of the brain and that they are particularly associated with neuronal function. Specific roles for ELOVL4 and its products in oligodendrocytes and myelin and in cellular proliferation, especially during development, are possible.
Keywords: cerebellum; cortex; hippocampus; seizure; spinocerebellar ataxia; stargardt-like macular dystrophy; very long chain fatty acids.
Figures








References
LinkOut - more resources
Full Text Sources
Other Literature Sources

