Intragenic DOK7 deletion detected by whole-genome sequencing in congenital myasthenic syndromes
- PMID: 28508085
- PMCID: PMC5415388
- DOI: 10.1212/NXG.0000000000000152
Intragenic DOK7 deletion detected by whole-genome sequencing in congenital myasthenic syndromes
Abstract
Objective: To identify the genetic cause in a patient affected by ptosis and exercise-induced muscle weakness and diagnosed with congenital myasthenic syndromes (CMS) using whole-genome sequencing (WGS).
Methods: Candidate gene screening and WGS analysis were performed in the case. Allele-specific PCR was subsequently performed to confirm the copy number variation (CNV) that was suspected from the WGS results.
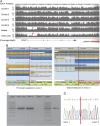
Results: In addition to the previously reported frameshift mutation c.1124_1127dup, an intragenic 6,261 bp deletion spanning from the 5' untranslated region to intron 2 of the DOK7 gene was identified by WGS in the patient with CMS. The heterozygous deletion was suspected based on reduced coverage on WGS and confirmed by allele-specific PCR. The breakpoints had microhomology and an inverted repeat, which may have led to the development of the deletion during DNA replication.
Conclusions: We report a CMS case with identification of the breakpoints of the intragenic DOK7 deletion using WGS analysis. This case illustrates that CNVs undetected by Sanger sequencing may be identified by WGS and highlights their relevance in the molecular diagnosis of a treatable neurologic condition such as CMS.
Figures

References
-
- Beeson D, Higuchi O, Palace J, et al. Dok-7 mutations underlie a neuromuscular junction synaptopathy. Science 2006;313:1975–1978. - PubMed
-
- Das AS, Agamanolis DP, Cohen BH. Use of next-generation sequencing as a diagnostic tool for congenital myasthenic syndrome. Pediatr Neurol 2014;51:717–720. - PubMed
-
- Garg N, Yiannikas C, Hardy TA, et al. Late presentations of congenital myasthenic syndromes: how many do we miss? Muscle Nerve 2016;54:721–727. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
