In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers
- PMID: 28508101
- PMCID: PMC5508036
- DOI: 10.1007/s00401-017-1725-7
In-depth clinico-pathological examination of RNA foci in a large cohort of C9ORF72 expansion carriers
Abstract
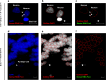
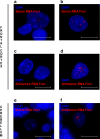
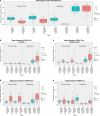
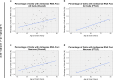
A growing body of evidence suggests that a loss of chromosome 9 open reading frame 72 (C9ORF72) expression, formation of dipeptide-repeat proteins, and generation of RNA foci contribute to disease pathogenesis in amyotrophic lateral sclerosis and frontotemporal dementia. Although the levels of C9ORF72 transcripts and dipeptide-repeat proteins have already been examined thoroughly, much remains unknown about the role of RNA foci in C9ORF72-linked diseases. As such, we performed a comprehensive RNA foci study in an extensive pathological cohort of C9ORF72 expansion carriers (n = 63). We evaluated two brain regions using a newly developed computer-automated pipeline allowing recognition of cell nuclei and RNA foci (sense and antisense) supplemented by manual counting. In the frontal cortex, the percentage of cells with sense or antisense RNA foci was 26 or 12%, respectively. In the cerebellum, 23% of granule cells contained sense RNA foci and 1% antisense RNA foci. Interestingly, the highest percentage of cells with RNA foci was observed in cerebellar Purkinje cells (~70%). In general, more cells contained sense RNA foci than antisense RNA foci; however, when antisense RNA foci were present, they were usually more abundant. We also observed that an increase in the percentage of cells with antisense RNA foci was associated with a delayed age at onset in the frontal cortex (r = 0.43, p = 0.003), whereas no other associations with clinico-pathological features were seen. Importantly, our large-scale study is the first to provide conclusive evidence that RNA foci are not the determining factor of the clinico-pathological variability observed in C9ORF72 expansion carriers and it emphasizes that the distribution of RNA foci does not follow the pattern of neurodegeneration, stressing the complex interplay between different aspects of C9ORF72-related diseases.
Keywords: Amyotrophic lateral sclerosis; C9ORF72; Frontotemporal dementia; Frontotemporal lobar degeneration; Motor neuron disease; RNA foci.
Conflict of interest statement
MDJ and RR hold a patent on methods to screen for the hexanucleotide repeat expansion in the
Figures



References
-
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. - DOI - PMC - PubMed
-
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. - DOI - PMC - PubMed
-
- Bieniek KF, Murray ME, Rutherford NJ, Castanedes-Casey M, DeJesus-Hernandez M, Liesinger AM, Baker MC, Boylan KB, Rademakers R, Dickson DW. Tau pathology in frontotemporal lobar degeneration with C9ORF72 hexanucleotide repeat expansion. Acta Neuropathol. 2013;125:289–302. doi: 10.1007/s00401-012-1048-7. - DOI - PMC - PubMed
-
- Cacace R, Van Cauwenberghe C, Bettens K, Gijselinck I, van der Zee J, Engelborghs S, Vandenbulcke M, Van Dongen J, Baumer V, Dillen L, et al. C9orf72 G4C2 repeat expansions in Alzheimer’s disease and mild cognitive impairment. Neurobiol Aging. 2013;34(1712):e1711–e1717. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

