Convergence of gut microbiotas in the adaptive radiations of African cichlid fishes
- PMID: 28509910
- PMCID: PMC5560477
- DOI: 10.1038/ismej.2017.62
Convergence of gut microbiotas in the adaptive radiations of African cichlid fishes
Abstract
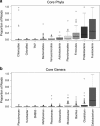
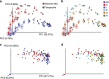
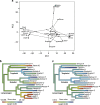
Ecoevolutionary dynamics of the gut microbiota at the macroscale level, that is, in across-species comparisons, are largely driven by ecological variables and host genotype. The repeated explosive radiations of African cichlid fishes in distinct lakes, following a dietary diversification in a context of reduced genetic diversity, provide a natural setup to explore convergence, divergence and repeatability in patterns of microbiota dynamics as a function of the host diet, phylogeny and environment. Here we characterized by 16S rRNA amplicon sequencing the gut microbiota of 29 cichlid species from two distinct lakes/radiations (Tanganyika and Barombi Mbo) and across a broad dietary and phylogenetic range. Within each lake, a significant deviation between a carnivorous and herbivorous lifestyle was found. Herbivore species were characterized by an increased bacterial taxonomic and functional diversity and converged in key compositional and functional community aspects. Despite a significant lake effect on the microbiota structure, this process has occurred with remarkable parallels in the two lakes. A metabolic signature most likely explains this trend, as indicated by a significant enrichment in herbivores/omnivores of bacterial taxa and functions associated with fiber degradation and detoxification of plant chemical compounds. Overall, compositional and functional aspects of the gut microbiota individually and altogether validate and predict main cichlid dietary habits, suggesting a fundamental role of gut bacteria in cichlid niche expansion and adaptation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Adams DC. (2014). A generalized K statistic for estimating phylogenetic signal from shape and other high-dimensional multivariate data. Syst Biol 63: 685–697. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

