Dynamic light scattering: a practical guide and applications in biomedical sciences
- PMID: 28510011
- PMCID: PMC5425802
- DOI: 10.1007/s12551-016-0218-6
Dynamic light scattering: a practical guide and applications in biomedical sciences
Abstract
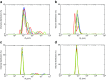
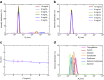
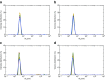
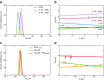
Dynamic light scattering (DLS), also known as photon correlation spectroscopy (PCS), is a very powerful tool for studying the diffusion behaviour of macromolecules in solution. The diffusion coefficient, and hence the hydrodynamic radii calculated from it, depends on the size and shape of macromolecules. In this review, we provide evidence of the usefulness of DLS to study the homogeneity of proteins, nucleic acids, and complexes of protein-protein or protein-nucleic acid preparations, as well as to study protein-small molecule interactions. Further, we provide examples of DLS's application both as a complementary method to analytical ultracentrifugation studies and as a screening tool to validate solution scattering models using determined hydrodynamic radii.
Keywords: Analytical ultracentrifuge; Diffusion coefficient; Dynamic light scattering; Hydrodynamic radius; Light scattering; Protein–ligand interactions; Protein–nucleic acid complexes; Protein–protein complexes.
Conflict of interest statement
Conflict of Interests
Jörg Stetefeld declares that he has no conflicts of interest. Sean A. McKenna declares that he has no conflicts of interest. Trushar R. Patel declares that he has no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures

References
-
- Amoros D, Ortega A, de la Torre JG. Hydrodynamic properties of wormlike macromolecules: Monte Carlo simulation and global analysis of experimental data. Macromolecules. 2011;44:5788–5797. doi: 10.1021/ma102697q. - DOI
-
- Barnett CE. Some applications of wave-length turbidimetry in the infrared. J Phys Chem. 1942;46:69–75. doi: 10.1021/j150415a009. - DOI
-
- Benitez AA, Hernandez Cifre JG, Diaz Banos FG, de la Torre JG. Prediction of solution properties and dynamics of RNAs by means of Brownian dynamics simulation of coarse-grained models: Ribosomal 5S RNA and phenylalanine transfer RNA. BMC Biophys. 2015;8:11. doi: 10.1186/s13628-015-0025-7. - DOI - PMC - PubMed
-
- Berne BJ, Pecora R. Dynamic light scattering: with applications to chemistry, biology, and physics. New York, USA: John Wiley & Sons, Inc; 1976.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

