Molecular assembly and structure of the bacteriophage T4 tail
- PMID: 28510021
- PMCID: PMC5418481
- DOI: 10.1007/s12551-016-0230-x
Molecular assembly and structure of the bacteriophage T4 tail
Abstract
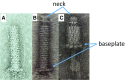
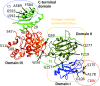
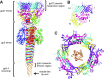
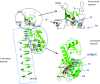
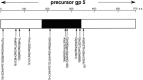
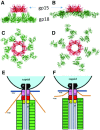
The tail of bacteriophage T4 undergoes large structural changes upon infection while delivering the phage genome into the host cell. The baseplate is located at the distal end of the contractile tail and plays a central role in transmitting the signal to the tail sheath that the tailfibers have been adsorbed by a host bacterium. This then triggers the sheath contraction. In order to understand the mechanism of assembly and conformational changes of the baseplate upon infection, we have determined the structure of an in vitro assembled baseplate through the three-dimensional reconstruction of cryo-electron microscopy images to a resolution of 3.8 Å from electron micrographs. The atomic structure was fitted to the baseplate structure before and after sheath contraction in order to elucidate the conformational changes that occur after bacteriophage T4 has attached itself to a cell surface. The structure was also used to investigate the protease digestion of the assembly intermediates and the mutation sites of the tail genes, resulting in a number of phenotypes.
Keywords: Assembly; Bacteriophage; Contractile tail; Infection; Molecular recognition; Tail baseplate.
Conflict of interest statement
Conflict of interest
Fumio Arisaka declares that none of the authors have any conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by the author.
Figures



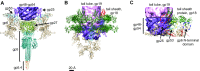

References
-
- Arisaka F, Engel J, Klump J. Contraction and dissociation of the bacteriophage T4 tail sheath induced by heat and urea. In: DuBow M, editor. Bacteriophage assembly. New York: Alan R. Liss; 1981. pp. 365–379. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

