Aspects of protein-DNA interactions: a review of quantitative thermodynamic theory for modelling synthetic circuits utilising LacI and CI repressors, IPTG and the reporter gene lacZ
- PMID: 28510022
- PMCID: PMC5425810
- DOI: 10.1007/s12551-016-0231-9
Aspects of protein-DNA interactions: a review of quantitative thermodynamic theory for modelling synthetic circuits utilising LacI and CI repressors, IPTG and the reporter gene lacZ
Abstract
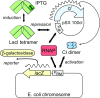
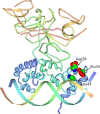
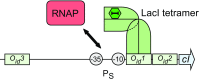
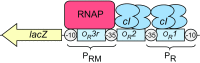
Protein-DNA interactions are central to the control of gene expression across all forms of life. The development of approaches to rigorously model such interactions has often been hindered both by a lack of quantitative binding data and by the difficulty in accounting for parameters relevant to the intracellular situation, such as DNA looping and thermodynamic non-ideality. Here, we review these considerations by developing a thermodynamically based mathematical model that attempts to simulate the functioning of an Escherichia coli expression system incorporating two of the best characterised prokaryotic DNA binding proteins, Lac repressor and lambda CI repressor. The key aim was to reproduce experimentally observed reporter gene activities arising from the expression of either wild-type CI repressor or one of three positive-control CI mutants. The model considers the role of several potentially important, but sometimes neglected, biochemical features, including DNA looping, macromolecular crowding and non-specific binding, and allowed us to obtain association constants for the binding of CI and its variants to a specific operator sequence.
Keywords: Escherichia coli expression system; Lambda CI repressor; Mathematical model; Synthetic biology; lac repressor.
Conflict of interest statement
Conflict of interest
Peter D. Munro declares that he has no conflict of interest. Gary K. Ackers declares that he has no conflict of interest. Keith E. Shearwin declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
References
-
- Bremer H, Dennis PP (1996) Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss R, Ingraham JL et al (eds) Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC, pp 1553–1569
-
- Chen J, Alberti S, Matthews KS. Wild-type operator binding and altered cooperativity for inducer binding of lac repressor dimer mutant R3. J Biol Chem. 1994;269:12482–12487. - PubMed
-
- Craven GR, Steers E, Anfinsen CB. Purification, composition, and molecular weight of the beta-galactosidase of Escherichia Coli K12. J Biol Chem. 1965;240:2468–2477. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

