New direct dynamic models of protein interactions coupled to photosynthetic electron transport reactions
- PMID: 28510068
- PMCID: PMC5425662
- DOI: 10.1007/s12551-010-0033-4
New direct dynamic models of protein interactions coupled to photosynthetic electron transport reactions
Abstract
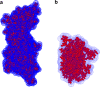
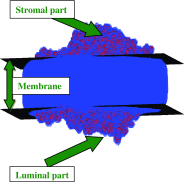
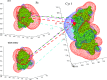
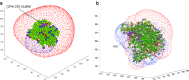
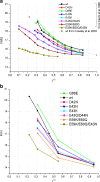
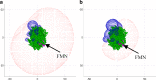
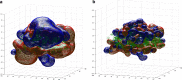
This review covers the methods of computer simulation of protein interactions taking part in photosynthetic electron transport reactions. A direct multiparticle simulation method that simulates reactions describing interactions of ensembles of molecules in the heterogeneous interior of a cell is developed. In the models, protein molecules move according to the laws of Brownian dynamics, mutually orient themselves in the electrical field, and form complexes in the 3D scene. The method allows us to visualize the processes of molecule interactions and to calculate the rate constants for protein complex formation reactions in the solution and in the photosynthetic membrane. Three-dimensional multiparticle computer models for simulating the complex formation kinetics for plastocyanin with photosystem I and cytochrome bf complex, and ferredoxin with photosystem I and ferredoxin:NADP+-reductase are considered. Effects of ionic strength are featured for wild type and mutant proteins. The computer multiparticle models describe nonmonotonic dependences of complex formation rates on the ionic strength as the result of long-range electrostatic interactions.
Keywords: Complex formation; Computer model; Electrostatic interaction; Photosynthesis; Protein interaction.
Figures






Similar articles
-
Molecular, Brownian, kinetic and stochastic models of the processes in photosynthetic membrane of green plants and microalgae.Biophys Rev. 2022 Aug 19;14(4):985-1004. doi: 10.1007/s12551-022-00988-w. eCollection 2022 Aug. Biophys Rev. 2022. PMID: 36124262 Free PMC article. Review.
-
[Multiparticle computer simulation of protein interactions in the photosynthetic membrane].Biofizika. 2011 Sep-Oct;56(5):775-86. Biofizika. 2011. PMID: 22117434 Russian.
-
Multiparticle Brownian dynamics simulation of experimental kinetics of cytochrome bf oxidation and photosystem I reduction by plastocyanin.Physiol Plant. 2017 Sep;161(1):88-96. doi: 10.1111/ppl.12570. Epub 2017 May 18. Physiol Plant. 2017. PMID: 28369912
-
Direct simulation of plastocyanin and cytochrome f interactions in solution.Phys Biol. 2006 Jun 5;3(2):121-9. doi: 10.1088/1478-3975/3/2/004. Phys Biol. 2006. PMID: 16829698
-
Electron transfers amongst cytochrome f, plastocyanin and photosystem I: kinetics and mechanisms.Biochim Biophys Acta. 2000 Jan 3;1456(1):5-26. doi: 10.1016/s0005-2728(99)00101-2. Biochim Biophys Acta. 2000. PMID: 10611452 Review.
Cited by
-
Molecular, Brownian, kinetic and stochastic models of the processes in photosynthetic membrane of green plants and microalgae.Biophys Rev. 2022 Aug 19;14(4):985-1004. doi: 10.1007/s12551-022-00988-w. eCollection 2022 Aug. Biophys Rev. 2022. PMID: 36124262 Free PMC article. Review.
-
Mechanisms of interaction of electron transport proteins in photosynthetic membranes of cyanobacteria.Dokl Biochem Biophys. 2011 Sep-Oct;440:213-5. doi: 10.1134/S160767291105005X. Epub 2011 Nov 19. Dokl Biochem Biophys. 2011. PMID: 22095121 No abstract available.
References
-
- Abaturova AM, Kovalenko IB, Riznichenko GY, Rubin AB. Direct multiparticle computer simulation of ionic strength dependence of the association rate constant for flavodoxin and photosystem I. Mathematics. Comput Educ. 2008;15(3):925–933.
-
- Belyaeva NE, Schmitt F-J, Steffen R, Paschenko VZ, Riznichenko GY, Chemeris YK, Renger G, Rubin AB. PSII model-based simulations of single turnover flash-induced transients of fluorescence yield monitored within the time domain of 100 ns–10 s on dark-adapted Chlorella pyrenoidosa cells. Photosynth Res. 2008;98:105–119. doi: 10.1007/s11120-008-9374-2. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

